Last updated on 2019-09-30 by Xiang-Jun Lu <xiangjun@x3dna.org>.
The block schematics were created with DSSR and
rendered using PyMOL.
- PDB-id
- 6ML6
- Class
- transcription-DNA
- Method
- X-ray (1.54 Å)
- Summary
- Zbtb24 zinc fingers 4-8 with 19+1mer DNA oligonucleotide (sequence 4 with a cpa 5mc modification)
List of 1 5mC-amino acid contact:
-
E.5CM4: stacking-with-A.ASN503 is-WC-paired is-in-duplex [+]:GcA/TGC
direct SNAP output · DNAproDB 2.0
- Reference
- Ren, R., Hardikar, S., Horton, J.R., Lu, Y., Zeng, Y., Singh, A.K., Lin, K., Coletta, L.D., Shen, J., Shuet, C., Kong, L., Hashimoto, H., Zhang, X., Chen, T., Cheng, X.: (2019) "Structural basis of specific DNA binding by the transcription factor ZBTB24." Nucleic Acids Res.
- Abstract
- ZBTB24, encoding a protein of the ZBTB family of transcriptional regulators, is one of four known genes-the other three being DNMT3B, CDCA7 and HELLS-that are mutated in immunodeficiency, centromeric instability and facial anomalies (ICF) syndrome, a genetic disorder characterized by DNA hypomethylation and antibody deficiency. The molecular mechanisms by which ZBTB24 regulates gene expression and the biological functions of ZBTB24 are poorly understood. Here, we identified a 12-bp consensus sequence [CT(G/T)CCAGGACCT] occupied by ZBTB24 in the mouse genome. The sequence is present at multiple loci, including the Cdca7 promoter region, and ZBTB24 binding is mostly associated with gene activation. Crystallography and DNA-binding data revealed that the last four of the eight zinc fingers (ZFs) (i.e. ZF5-8) in ZBTB24 confer specificity of DNA binding. Two ICF missense mutations have been identified in the ZBTB24 ZF domain, which alter zinc-binding cysteine residues. We demonstrated that the corresponding C382Y and C407G mutations in mouse ZBTB24 abolish specific DNA binding and fail to induce Cdca7 expression. Our analyses indicate and suggest a structural basis for the sequence specific recognition by a transcription factor centrally important for the pathogenesis of ICF syndrome.
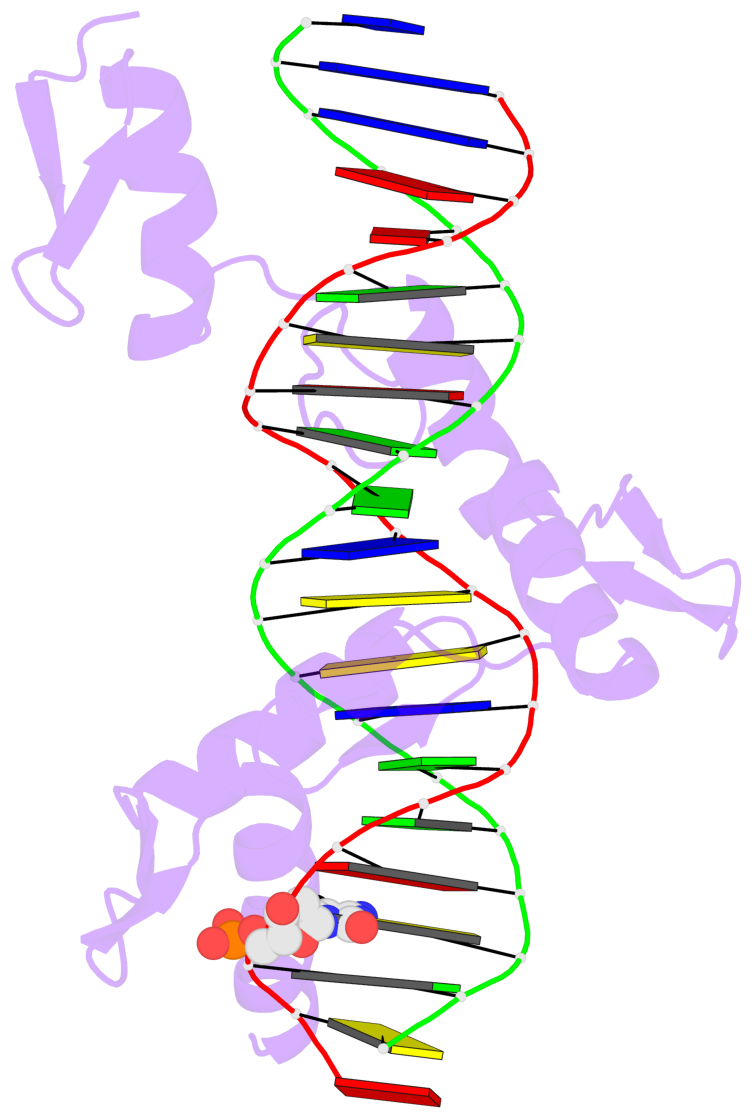
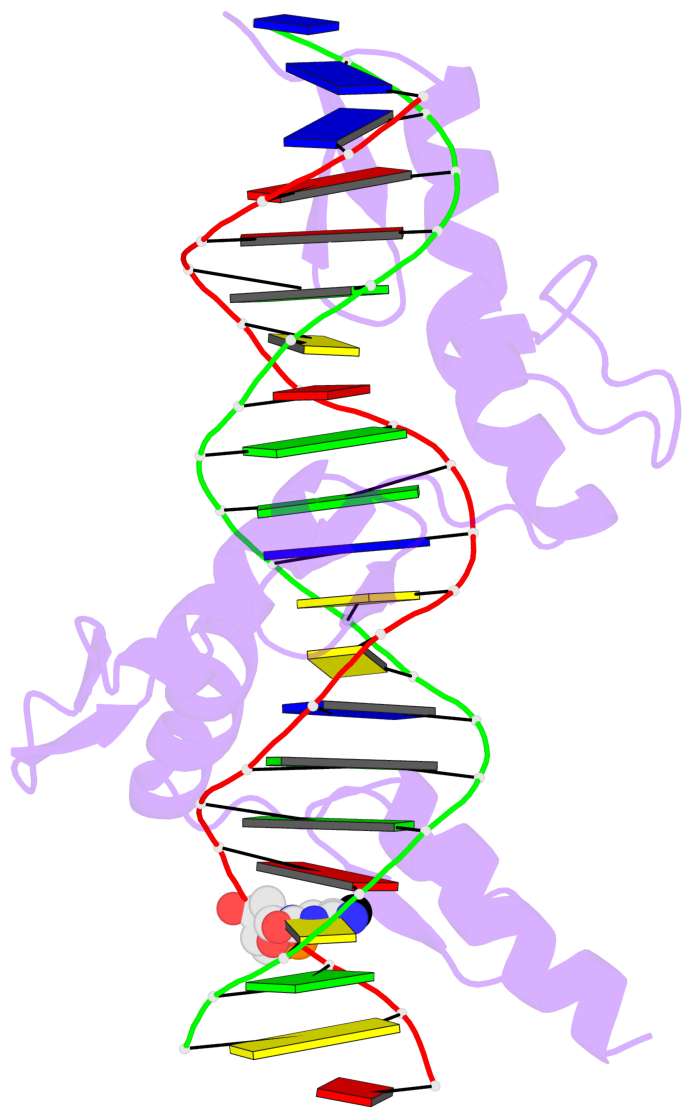
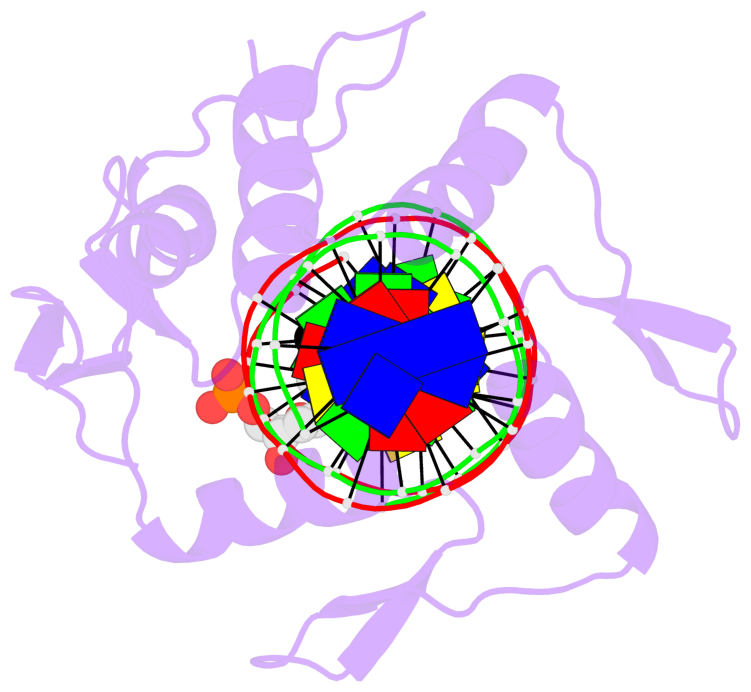
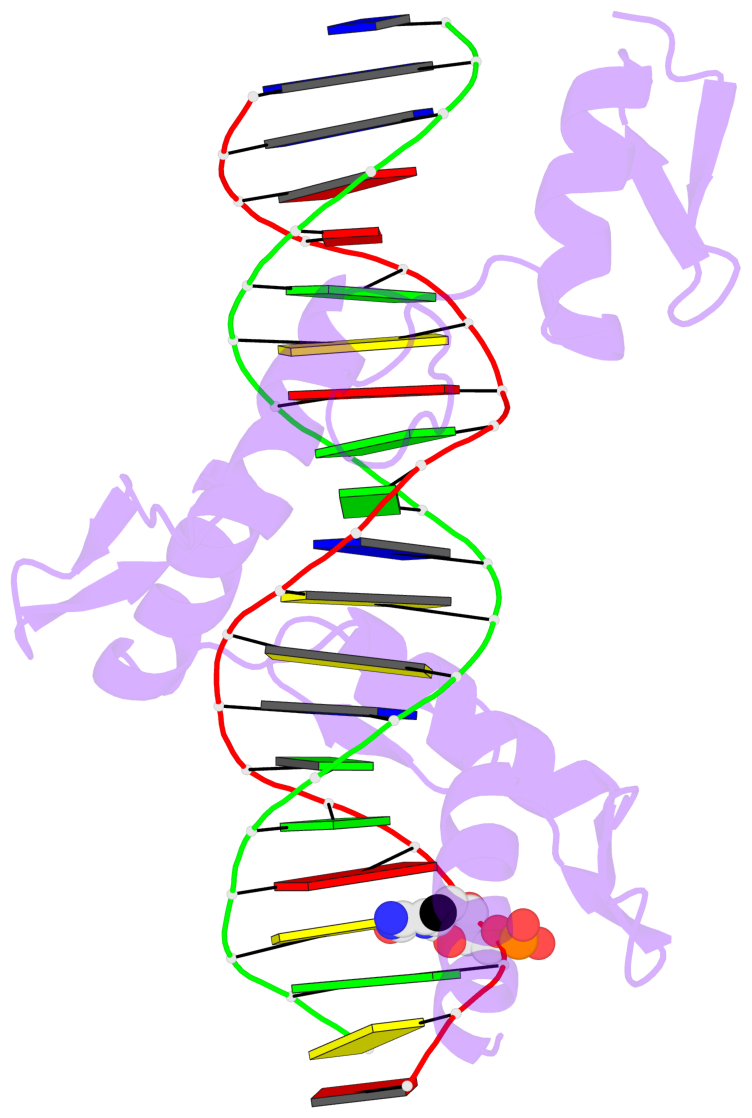
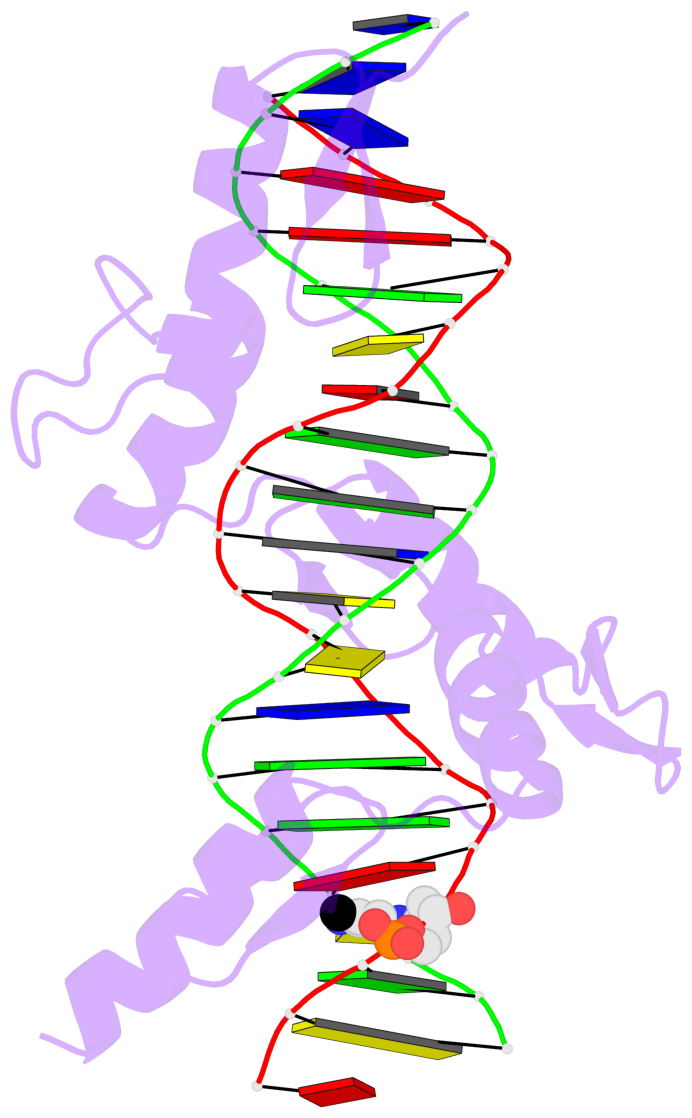
- The 5-methylcytosine group (PDB ligand '5CM') is shown in space-filling model,
with the methyl-carbon atom in black.
- Watson-Crick base pairs are represented as long rectangular blocks with the
minor-groove edge in black. Color code: A-T red, C-G yellow, G-C green, T-A blue.
- Protein is shown as cartoon in purple. DNA backbones are shown ribbon, colored code
by chain identifier.
- The block schematics were created with 3DNA-DSSR,
and images were rendered using PyMOL.
- Download the PyMOL session file corresponding to the top-left
image in the following panel.
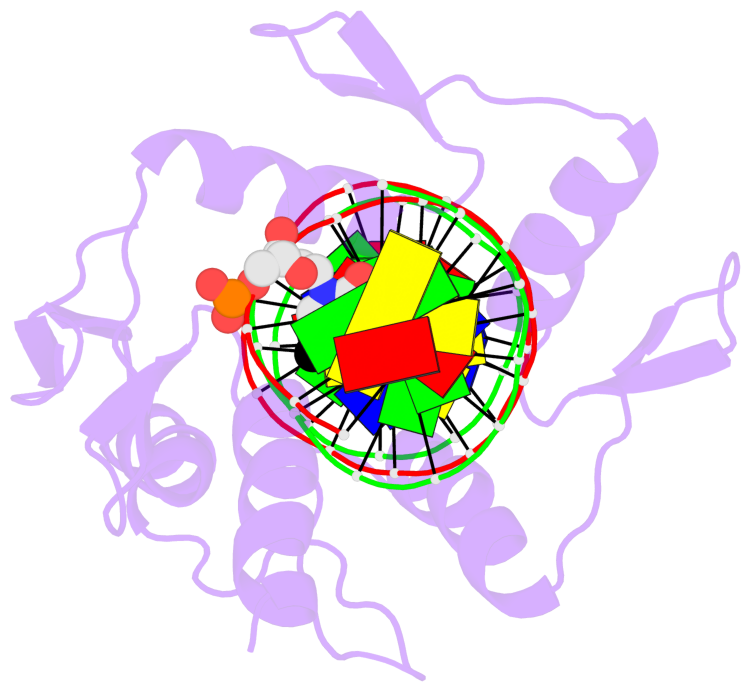
- The contacts include paired nucleotides (mostly a G in G-C pairing), and
amino-acids within a 4.5-A distance cutoff to the base atoms of 5mC.
- The structure is oriented in the 'standard' base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
 |
No. 1 E.5CM4: download PDB file
for the 5mC entry
stacking-with-A.ASN503 is-WC-paired is-in-duplex [+]:GcA/TGC
|