Tutorial on SNAP-enabled auto-curation of PDB TF-DNA complexes containing 5-methyl-cytosines
Selection of protein-DNA complexes in the PDB
Search criteria
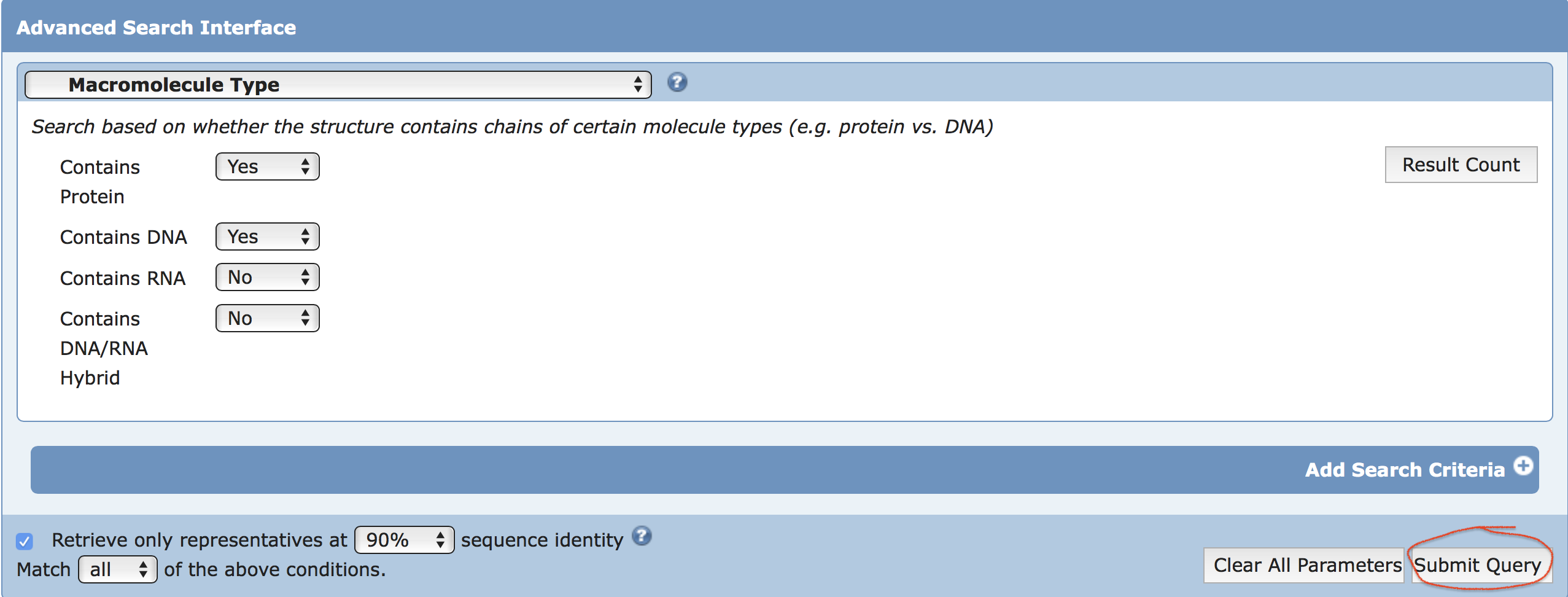
A search was performed on 2019-09-28 using RCSB PDB with the following criteria (see the screenshot):
Chain Type: there is a Protein and a DNA chain but not any
RNA or Hybrid and Representative Structures at 90% Sequence
Identity
Summary report in CSV format
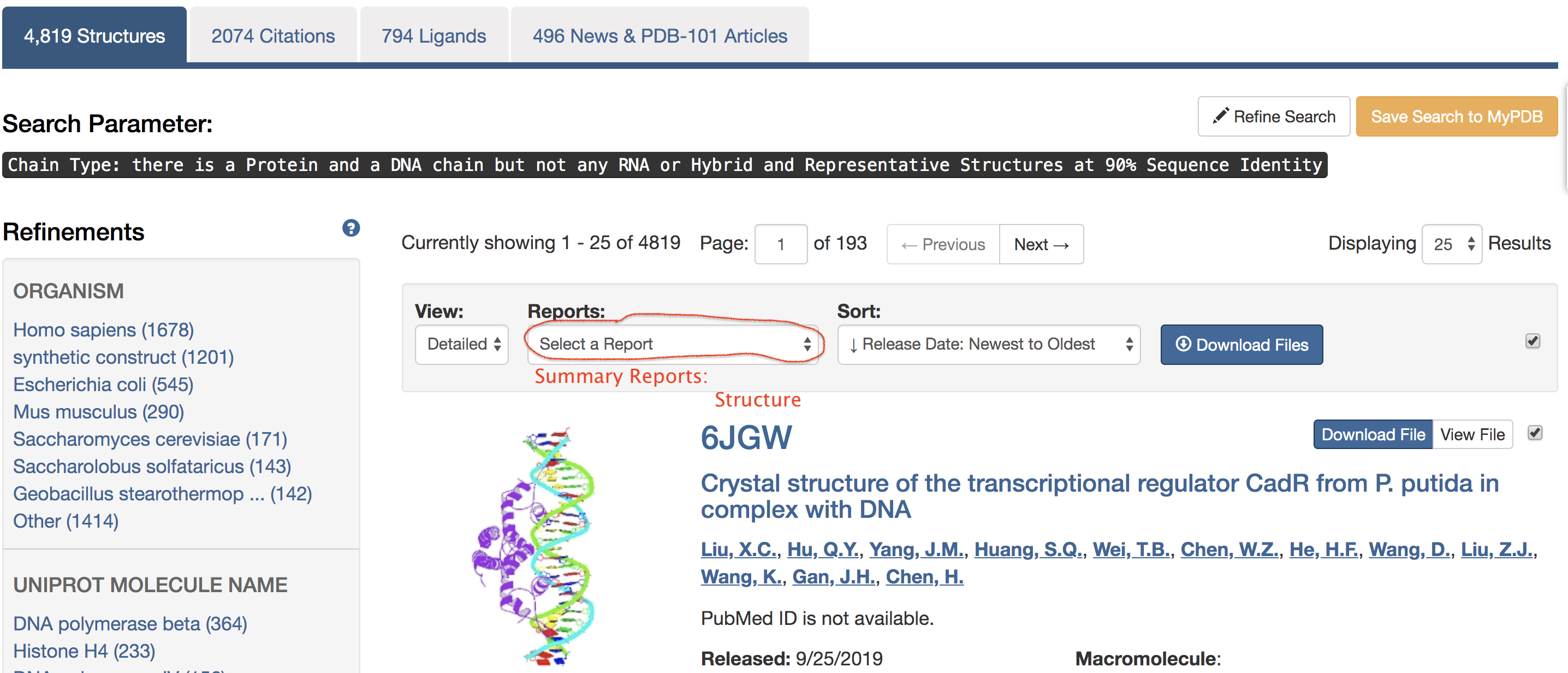
Clicking the "Submit Query" button led to the result page,
showing a total of 4,819 hits under the search
criteria. Then click on the "Reports" drop-down list, and
select the "Structure" option under the section of "Summary
Reports", as shown in the screenshot below:
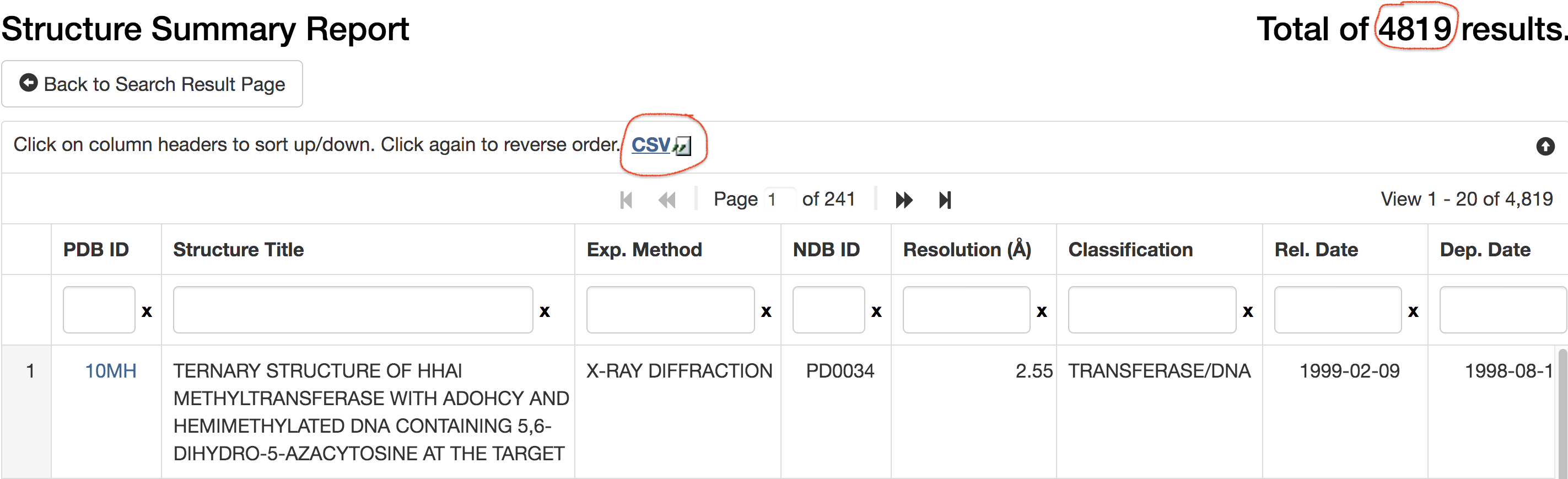
The "Structure Summary Report" page then appeared. Save the
result of the 4,819 hits in CSV format in a file named
"proDNA-2019sep26-summary.csv"
(see the screenshot below). The summary CSV contains the following
information (including PDB id), which forms the starting point of
later on SNAP analyses.
PDB ID, Structure Title, Exp. Method, NDB ID, Resolution,
Classification, Rel. Date, Dep. Date, Rev. Date, Structure Author,
Structure MW, Macromolecule Type, Residue Count, Atom Site Count,
PDB DOI
Identification and characterization of 5mC-DNA/protein interactions using SNAP
Download PDB coordinates files
With PDB id, the atomic coordinates file for each of the
4,819 DNA-protein complexes can be downloaded from the
RCSB PDB. In the following sections, PDB entry 4m9e
will be used as an example, whose coordinates file
(4m9e.pdb) was retrieved via the command:
wget https://files.rcsb.org/download/4m9e.pdb
Run SNAP on PDB coordinates files
SNAP is a general-purpose program, with many command-line
options, for characterizing DNA/RNA-protein interactions. Running
SNAP on PDB entry 4m9e using the default options is as
simple as specifying the input (4m9e.pdb) and output
file (4m9e.out):
x3dna-snap -i=4m9e.pdb -o=4m9e.out
The default SNAP output file 4m9e.out includes a
list of 2 modified nucleotides, 1 double helix, 48
nucleotide/amino-acid interactions, 40 base-pair/amino-acid
interactions, 4 phosphate/amino-acid H-bonds, 13 base/amino-acid
H-bonds, 8 base/amino-acid pairs, and 2 base/amino-acid stacks.
The list of the 2 base/amino-acid stacking interactions is shown below. One is 5CM (5-methylcytosine on chain B, residue number 5) with arginine#443 (on chain A), and the other is T7 over histidine#416.
List of 2 base/amino-acid stacks
id nt-aa nt aa vertical-distance plane-angle
1 4m9e C-arg B.5CM5 A.ARG443 3.40 5
2 4m9e T-his B.DT7 A.HIS416 3.33 11
The SNAP --methyl-C option
For an automatic analysis of
5-methyl-cytosine-containing TF-DNA complexes, the
--methyl-C (short-form:
--methyl) option was introduced into SNAP
(and documented as of v1.0.6-2019sep30). Note that per
PDB, 5-methylcytosine (5mC) in DNA is designated
5CM and the 5-methyl carbon atom is named
C5A (see
"5CM and 5MC, two forms of 5-methylcytosine in the
PDB"). Moreover, the --type=base
option is employed to ensure that base atoms
(regardless sugar-phosphate atoms) of 5mC are directly
involved in interactions with amino acids. So the SNAP
command becomes:
x3dna-snap --methyl-C
--type=base -i=4m9e.pdb -o=4m9e-5mC.out
With the SNAP --methyl option, two additional files
are also generated: a text file 4m9e-5mC.txt (as shown below) and a
corresponding PDB file 4m9e-5mC.pdb
(potentially multi-model, two as in this case).
4m9e:B.5CM5: stacking-with-A.ARG443 is-WC-paired is-in-duplex [+]:GcG/cGC 4m9e:C.5CM5: other-contacts is-WC-paired is-in-duplex [-]:cGT/AcG
For a DNA-protein complex that has no 5mC (e.g., PDB entry
1oct) or without 5mC-amino acid interactions (e.g.,
1odg), SNAP will not generate these two additional
files. Thus, output of these two files indicates the existence of 5mC
interacting with amino acids. Running SNAP with the --methyl-C
--type=base options on the 4,819 DNA-protein
complexes, we identified 107 entries with 5mC whose base
atoms interact with amino acids. The list of the 107
PDB entries is as below:
10mh, 1bsu, 1dct, 1ig4, 2c7o, 2c7p, 2c7q, 2c7r, 2ky8, 2moe,
2uyc, 2uyh, 2uz4, 2zkd, 2zke, 2zkf, 2zo0, 2zo1, 3c2i, 3clz, 3f8i,
3f8j, 3fde, 3q0b, 3q0c, 3q0d, 3q0f, 3ssc, 3ssd, 3vxv, 3vxx, 3vyb,
3vyq, 4aqu, 4aqx, 4da4, 4dkj, 4f6n, 4gjp, 4gjr, 4gzn, 4hp1, 4lg7,
4lt5, 4m9e, 4m9v, 4mht, 4nm6, 4pw7, 4qen, 4qeo, 4qep, 4r28, 4r2a,
4r2e, 4r2r, 4r2s, 4x9j, 5bt2, 5cg9, 5cpj, 5cpk, 5ef6, 5ego, 5emc,
5gse, 5ke7, 5ke8, 5kl4, 5kl5, 5kl7, 5lty, 5lux, 5mcv, 5mcw, 5mht,
5szx, 5t00, 5t01, 5vmu, 5vmv, 5vmw, 5vmx, 5vmy, 5vmz, 6a5n, 6c1a,
6c1t, 6c1u, 6c1y, 6c2f, 6cc8, 6ccg, 6ceu, 6cnp, 6cnq, 6d1t, 6e93,
6e94, 6jnm, 6jnn, 6mg2, 6mg3, 6mht, 6ml6, 6ml7, 6ogk
Among the the 107 PDB entries, 41 are
enzymes (e.g., ligase, transferase etc.), two are
solved by NMR (1ig4 and
2ky8), 4 x-ray crystal structures have
worse than 3.0-Å resolution (4qep,
5cpj, 5gse and
5lux). We thus have a total of 60
transcription factor-DNA complexes, as shown
below:
3c2i, 3ssc, 3ssd, 4f6n, 4gjp, 4gjr, 4gzn, 4hp1, 4m9e, 4m9v,
4qen, 4qeo, 4r2a, 4r2e, 4r2r, 4r2s, 4x9j, 5bt2, 5cpk, 5ef6, 5ego,
5emc, 5ke7, 5ke8, 5kl4, 5kl5, 5kl7, 5lty, 5mcv, 5mcw, 5szx, 5t00,
5t01, 5vmu, 5vmv, 5vmw, 5vmx, 5vmy, 5vmz, 6a5n, 6c1a, 6c1t, 6c1u,
6c1y, 6c2f, 6cc8, 6ccg, 6ceu, 6cnp, 6cnq, 6d1t, 6e93, 6e94, 6jnm,
6jnn, 6mg2, 6mg3, 6ml6, 6ml7, 6ogk
Three modes of 5mC recognition by TF
In this work, the 5mC recognition by a TF is classified into three
modes: (1) stacking interactions with a planar amino-acid side chain,
most notably arginine, (2) hydrophobic interactions with ILE, VAL,
ALA, MET, LEU, PRO, GLY, and (3) other than these two modes. As
shown below, for PDB entry 4m9e,
modes (1) and (3) are detected, whilst for 4gjr,
mode (2) is also available.
4m9e:B.5CM5: stacking-with-A.ARG443 is-WC-paired is-in-duplex [+]:GcG/cGC 4m9e:C.5CM5: other-contacts is-WC-paired is-in-duplex [-]:cGT/AcG 4gjr:I.5CM8: hydrophobic-with-A.GLY437 hydrophobic-with-A.GLY438 is-WC-paired is-in-duplex [+]:TcT/AGA
In the SNAP output, "is-WC-paired" means the 5mC is in a
Watson-Crick base pair, and "is-in-duplex" indicates that the base
pair is in a DNA double helix. The last portion shows the sequence
context of 5mC (abbreviated to lower-case 'c') with its immediate 5'
and 3' neighbors: [+] means the leading strand, whilst
[-] signifies the reverse complementary strand.
Bibliographic information
For the 60 entries with TF-5mC DNA interactions, we
extracted primary citations from the PDB, with
those fields: structureId, classification, structureTitle,
experimentalTechnique, resolution, title, citationAuthor,
journalName, publicationYear, volumeId, firstPage, lastPage,
pubmedId. When pubmedId is available for a PDB
entry, we downloaded the abstract from PubMED. The bibliographic
information and SNAP annotations are presented in a dynamic
table using DataTables.
Molecular visualization using DSSR, PyMOL and 3Dmol.js
Each of the TF-5mC PDB entries also has a detailed annotation webpage, with bibliographic information and molecular images.
DSSR-PyMOL block images
For the PDB entry as a whole, the DSSR-PyMOL schematic block images in
six orthogonal views (front, right, top; back, left, bottom) are
generated. Specifically, the DSSR command options used are:
--blocview --block-file=wc-minor to orient the
structure in the most extended view, and to show WC pairs as a long
block with the minor-groove edge colored in black. The PyMOL session
file corresponding to the top-left image is available for download
so users can reproduce the molecular images
rigorously.
Please visit the
'schematics' section in the SNAP-annotation page for PDB
entry 4m9e for an example.
Molecular images for 5mC interactions
Each 5mC is further zoomed-in, with bases and amino acids that
interacts with it. Moreover, the cluster of interacting residues is
oriented in the standard base reference frame of 5mC, allowing for
easy comparison and direct overlap of multiple clusters. The atomic
coordinates are transformed directly by SNAP, and stored in files
like 4m9e-5mC.pdb as noted above.
The static image is enabled by DSSR with options
--block-file=fill-hbond --cartoon-block=sticks-label
and rendered in PyMOL. It has filled base rings and H-bonds, and the
residues are shown in sticks and labeled. Moreover, a 3Dmol.js viewer is embedded for
interactive visualization.
Please visit the
'contacts' section in the SNAP-annotation page for PDB
entry 4m9e for an example.
Last updated on 2019-09-30 by Xiang-Jun Lu <xiangjun@x3dna.org>