Last updated on 2019-09-30 by Xiang-Jun Lu <xiangjun@x3dna.org>.
The block schematics were created with DSSR and
rendered using PyMOL.
- PDB-id
- 6MG2
- Class
- transcription-DNA
- Method
- X-ray (1.93 Å)
- Summary
- C-terminal bzip domain of human c-ebpbeta with 16bp methylated oligonucleotide containing consensus recognition sequence-c2221 crystal form
List of 4 5mC-amino acid contacts:
-
C.5CM8: stacking-with-A.ARG289 is-WC-paired is-in-duplex [+]:GcG/cGc
-
C.5CM10: hydrophobic-with-A.VAL285 is-WC-paired is-in-duplex [+]:GcA/TGc
-
D.5CM108: other-contacts is-WC-paired is-in-duplex [-]:cGc/GcG
-
D.5CM110: hydrophobic-with-B.VAL285 is-WC-paired is-in-duplex [-]:TGc/GcA
direct SNAP output · DNAproDB 2.0
- Reference
- Yang, J., Horton, J.R., Wang, D., Ren, R., Li, J., Sun, D., Huang, Y., Zhang, X., Blumenthal, R.M., Cheng, X.: (2019) "Structural basis for effects of CpA modifications on C/EBP beta binding of DNA." Nucleic Acids Res., 47, 1774-1785.
- Abstract
- CCAAT/enhancer binding proteins (C/EBPs) regulate gene expression in a variety of cells/tissues/organs, during a range of developmental stages, under both physiological and pathological conditions. C/EBP-related transcription factors have a consensus binding specificity of 5'-TTG-CG-CAA-3', with a central CpG/CpG and two outer CpA/TpG dinucleotides. Methylation of the CpG and CpA sites generates a DNA element with every pyrimidine having a methyl group in the 5-carbon position (thymine or 5-methylcytosine (5mC)). To understand the effects of both CpG and CpA modification on a centrally-important transcription factor, we show that C/EBPβ binds the methylated 8-bp element with modestly-increased (2.4-fold) binding affinity relative to the unmodified cognate sequence, while cytosine hydroxymethylation (particularly at the CpA sites) substantially decreased binding affinity (36-fold). The structure of C/EBPβ DNA binding domain in complex with methylated DNA revealed that the methyl groups of the 5mCpA/TpG make van der Waals contacts with Val285 in C/EBPβ. Arg289 recognizes the central 5mCpG by forming a methyl-Arg-G triad, and its conformation is constrained by Val285 and the 5mCpG methyl group. We substituted Val285 with Ala (V285A) in an Ala-Val dipeptide, to mimic the conserved Ala-Ala in many members of the basic leucine-zipper family of transcription factors, important in gene regulation, cell proliferation and oncogenesis. The V285A variant demonstrated a 90-fold binding preference for methylated DNA (particularly 5mCpA methylation) over the unmodified sequence. The smaller side chain of Ala285 permits Arg289 to adopt two alternative conformations, to interact in a similar fashion with either the central 5mCpG or the TpG of the opposite strand. Significantly, the best-studied cis-regulatory elements in RNA polymerase II promoters and enhancers have variable sequences corresponding to the central CpG or reduced to a single G:C base pair, but retain a conserved outer CpA sequence. Our analyses suggest an important modification-dependent CpA recognition by basic leucine-zipper transcription factors.
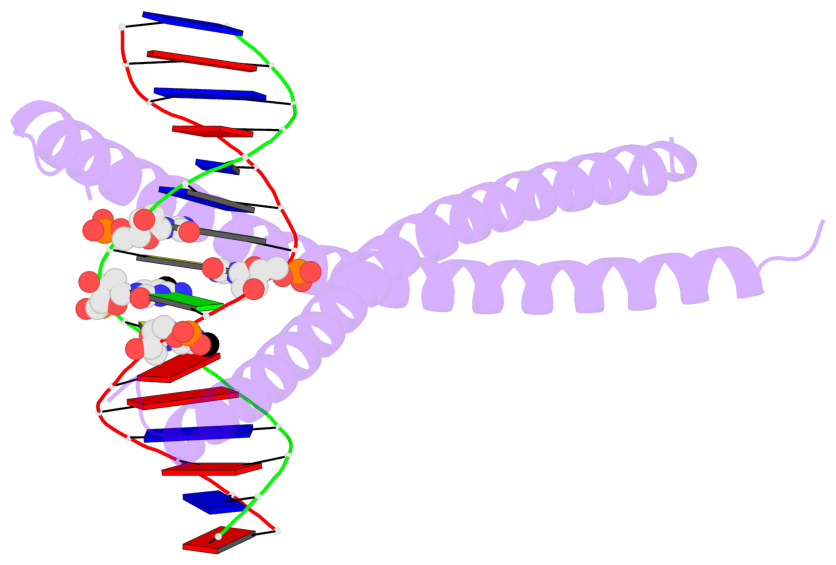
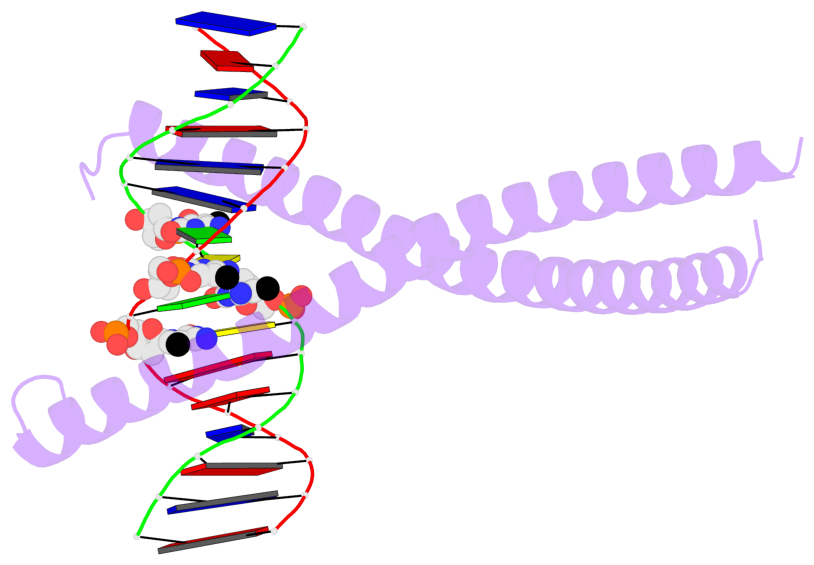
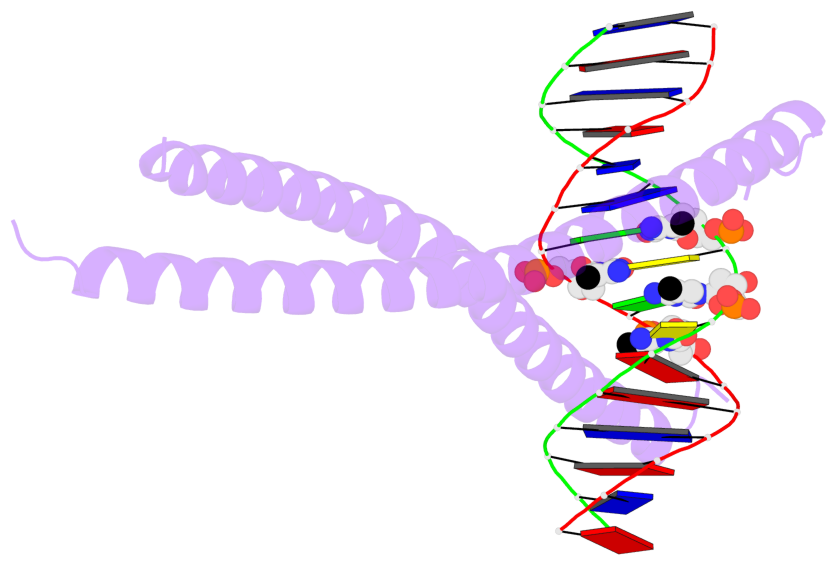
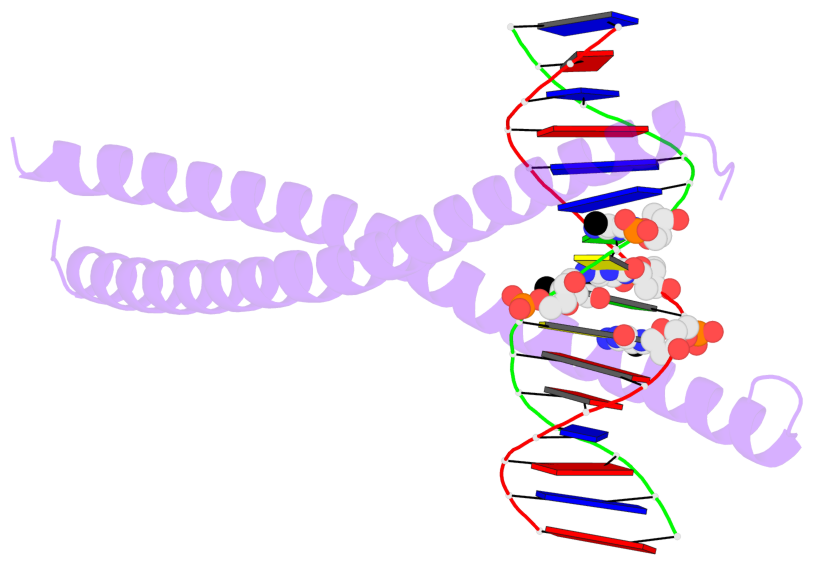
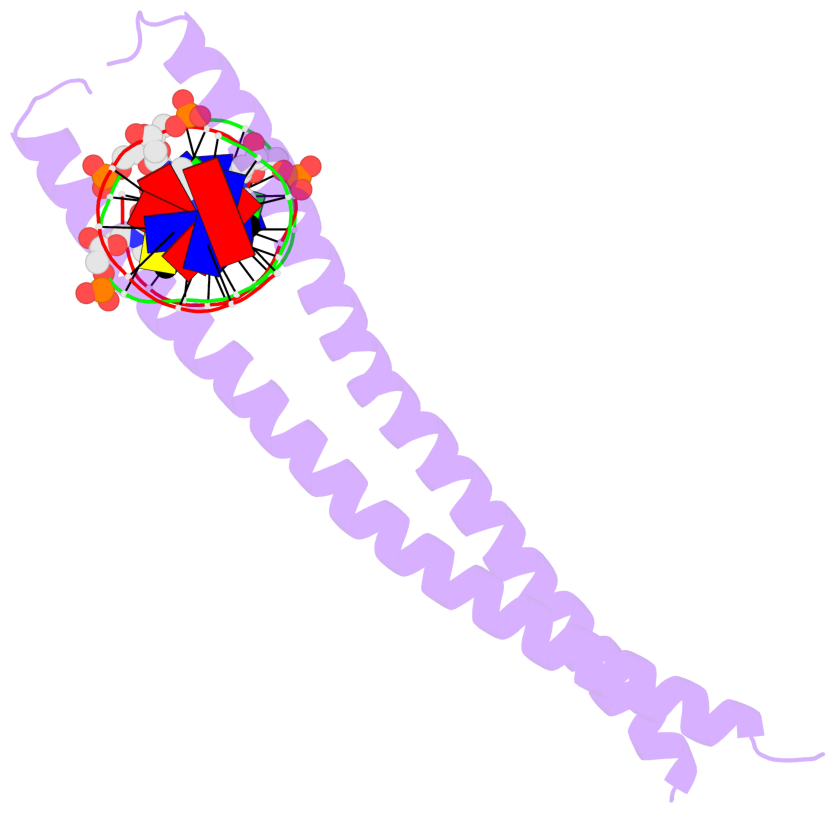
- The 5-methylcytosine group (PDB ligand '5CM') is shown in space-filling model,
with the methyl-carbon atom in black.
- Watson-Crick base pairs are represented as long rectangular blocks with the
minor-groove edge in black. Color code: A-T red, C-G yellow, G-C green, T-A blue.
- Protein is shown as cartoon in purple. DNA backbones are shown ribbon, colored code
by chain identifier.
- The block schematics were created with 3DNA-DSSR,
and images were rendered using PyMOL.
- Download the PyMOL session file corresponding to the top-left
image in the following panel.
- The contacts include paired nucleotides (mostly a G in G-C pairing), and
amino-acids within a 4.5-A distance cutoff to the base atoms of 5mC.
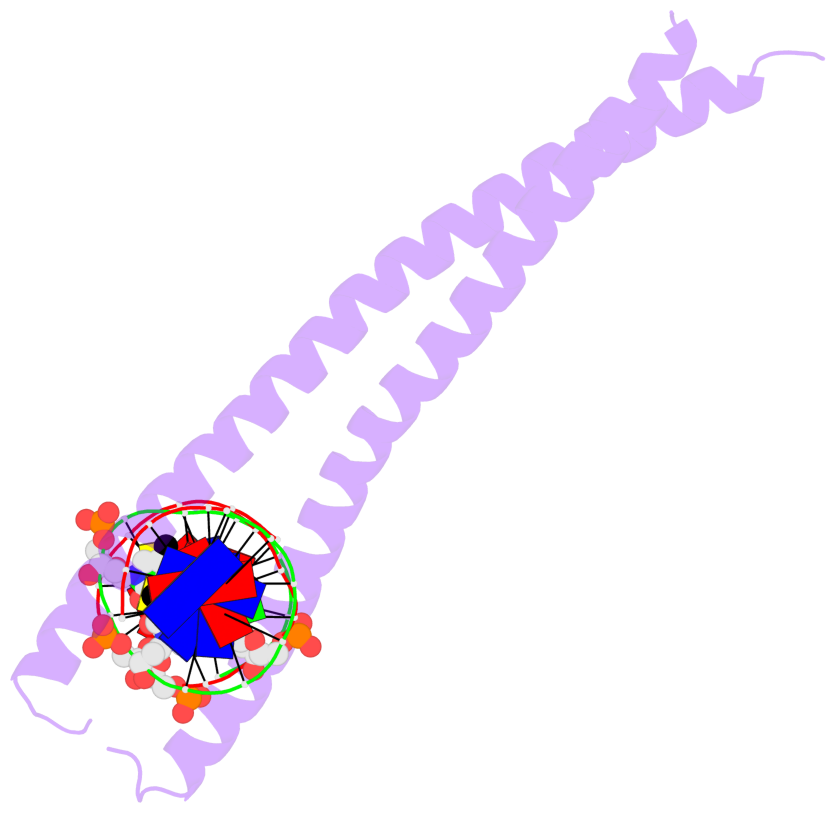
- The structure is oriented in the 'standard' base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
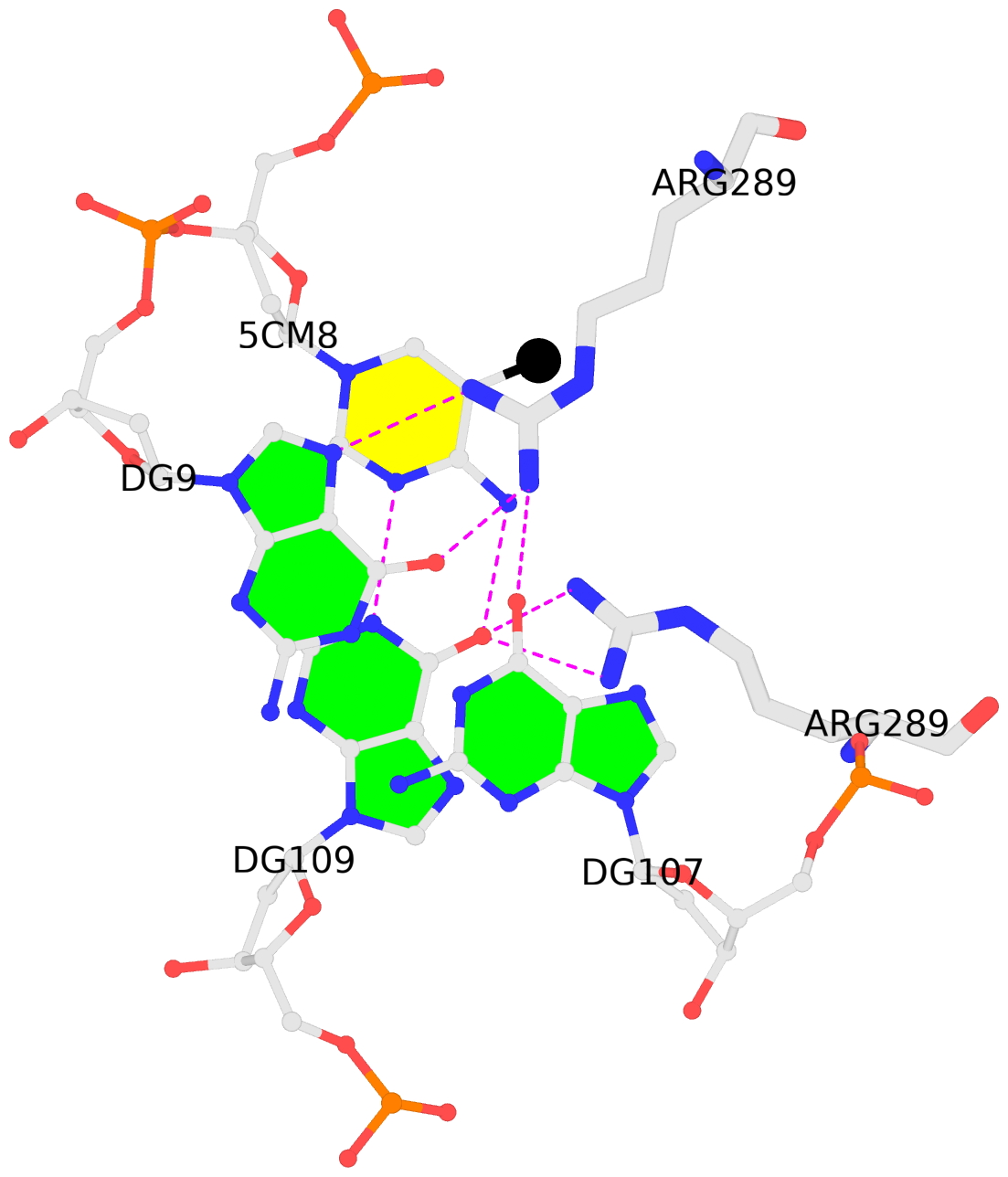 |
No. 1 C.5CM8: download PDB file
for the 5mC entry
stacking-with-A.ARG289 is-WC-paired is-in-duplex [+]:GcG/cGc
|
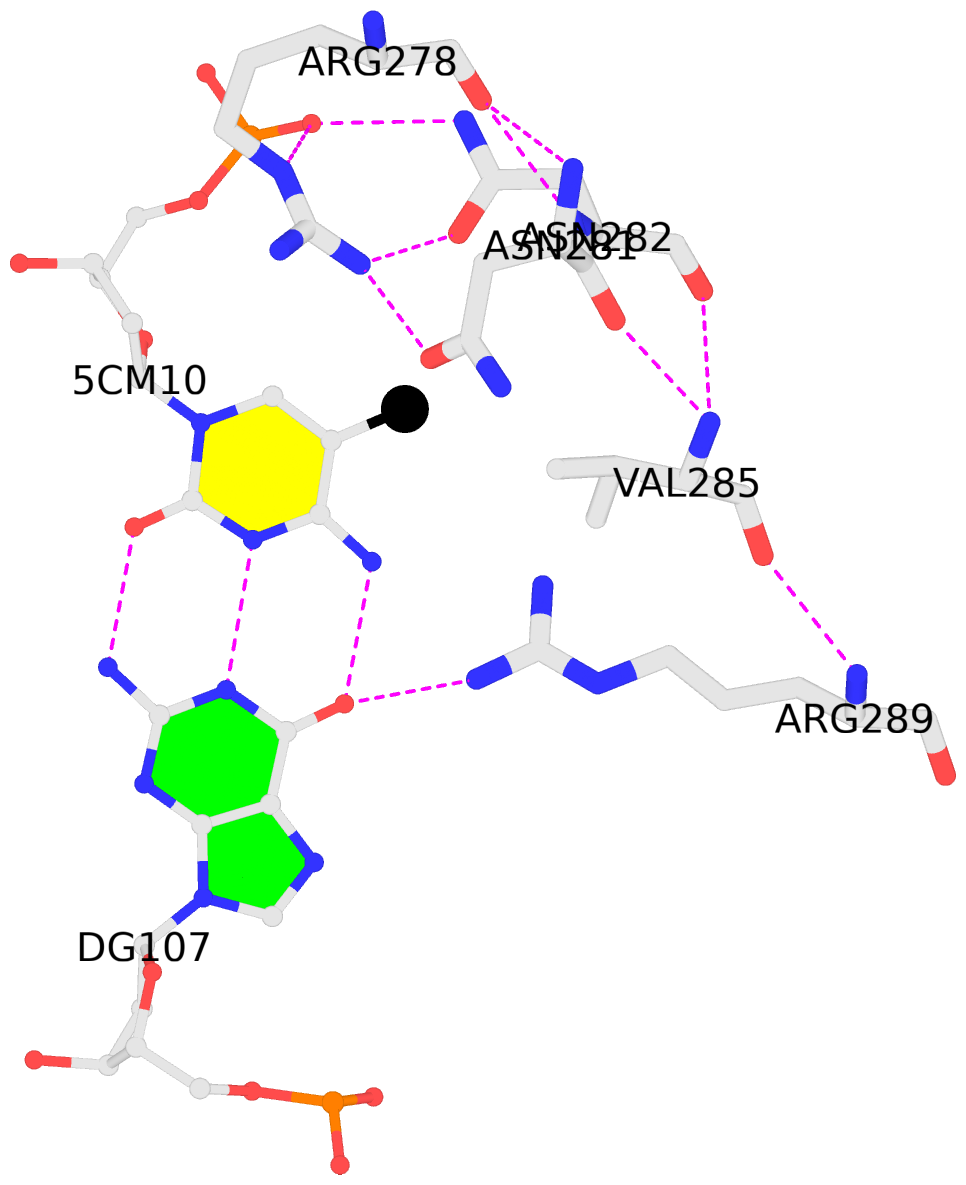 |
No. 2 C.5CM10: download PDB file
for the 5mC entry
hydrophobic-with-A.VAL285 is-WC-paired is-in-duplex [+]:GcA/TGc
|
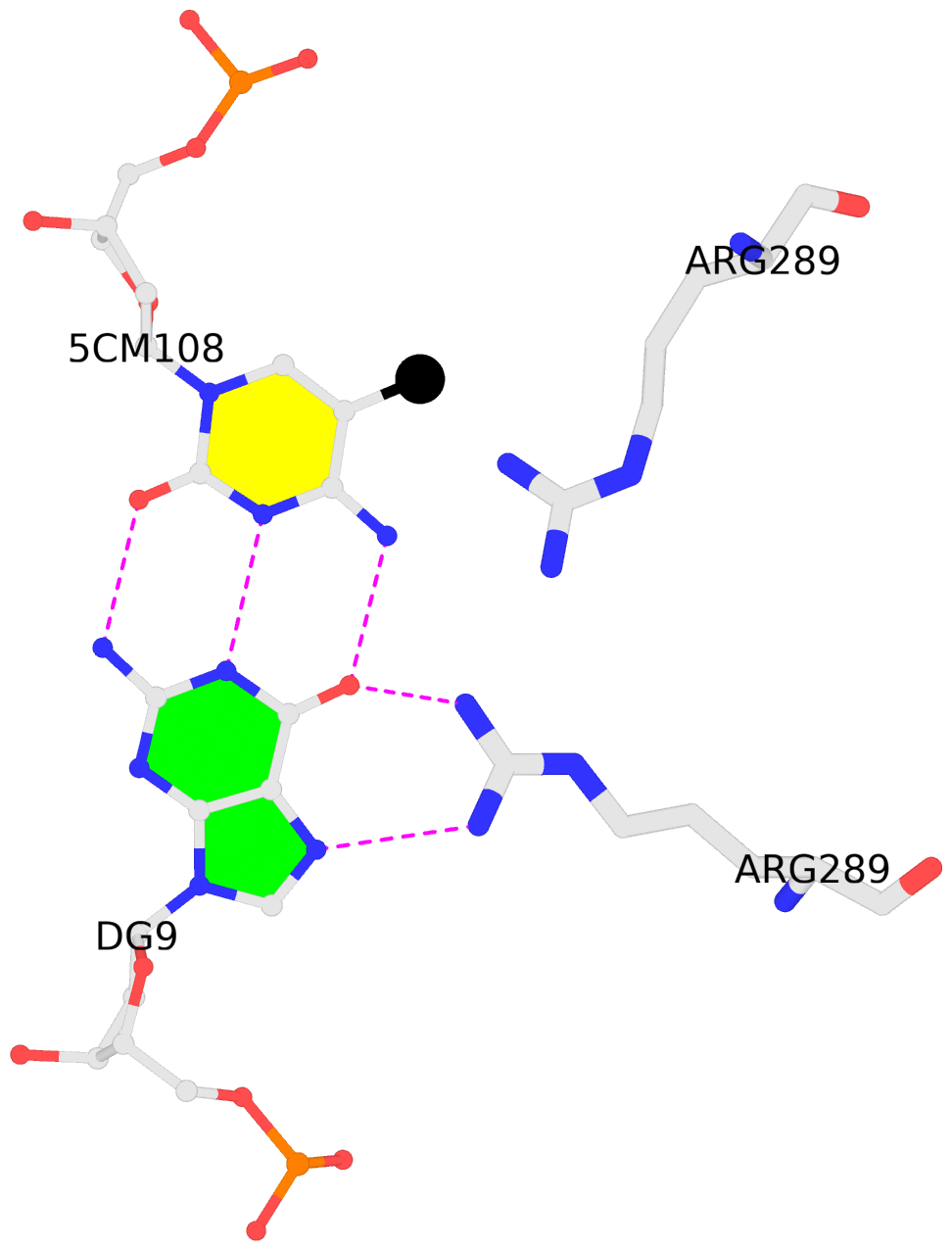 |
No. 3 D.5CM108: download PDB file
for the 5mC entry
other-contacts is-WC-paired is-in-duplex [-]:cGc/GcG
|
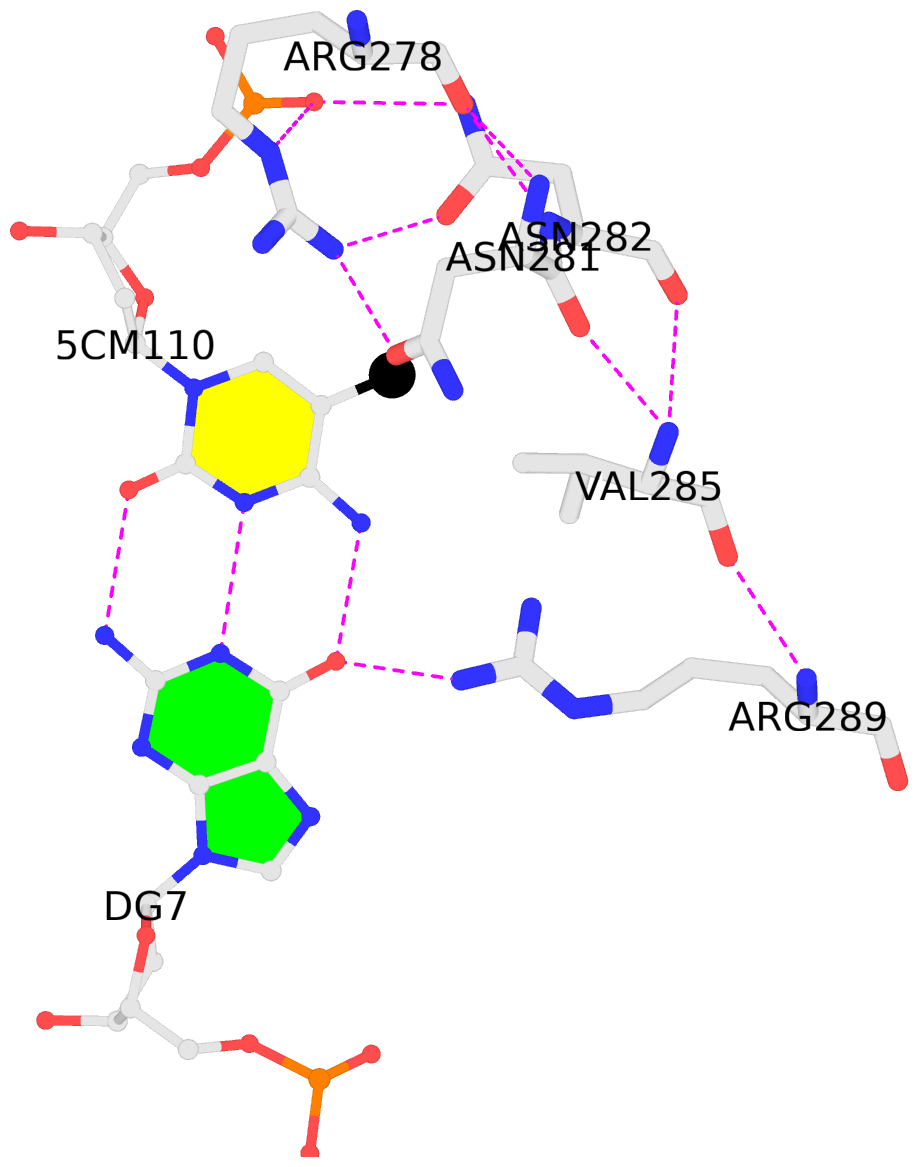 |
No. 4 D.5CM110: download PDB file
for the 5mC entry
hydrophobic-with-B.VAL285 is-WC-paired is-in-duplex [-]:TGc/GcA
|