Last updated on 2019-09-30 by Xiang-Jun Lu <xiangjun@x3dna.org>.
The block schematics were created with DSSR and
rendered using PyMOL.
- PDB-id
- 6JNN
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.6 Å)
- Summary
- Ref6 znf2-4-nac004-mc1 complex
List of 4 5mC-amino acid contacts:
-
D.5CM3: other-contacts is-WC-paired is-in-duplex [-]:AGA/TcT
-
F.5CM3: other-contacts is-WC-paired is-in-duplex [+]:TcT/AGA
-
I.5CM3: other-contacts is-WC-paired is-in-duplex [+]:TcT/AGA
-
L.5CM3: other-contacts is-WC-paired is-in-duplex [+]:TcT/AGA
direct SNAP output · DNAproDB 2.0
- Reference
- Qiu, Q., Mei, H., Deng, X., He, K., Wu, B., Yao, Q., Zhang, J., Lu, F., Ma, J., Cao, X.: (2019) "DNA methylation repels targeting of Arabidopsis REF6." Nat Commun, 10, 2063-2063.
- Abstract
- RELATIVE OF EARLY FLOWERING 6 (REF6/JMJ12), a Jumonji C (JmjC)-domain-containing H3K27me3 histone demethylase, finds its target loci in Arabidopsis genome by directly recognizing the CTCTGYTY motif via its zinc-finger (ZnF) domains. REF6 tends to bind motifs located in active chromatin states that are depleted for heterochromatic modifications. However, the underlying mechanism remains unknown. Here, we show that REF6 preferentially bind to hypo-methylated CTCTGYTY motifs in vivo, and that CHG methylation decreases REF6 DNA binding affinity in vitro. In addition, crystal structures of ZnF-clusters in complex with DNA oligonucleotides reveal that 5-methylcytosine is unfavorable for REF6 binding. In drm1 drm2 cmt2 cmt3 (ddcc) quadruple mutants, in which non-CG methylation is significantly reduced, REF6 can ectopically bind a small number of new target loci, most of which are located in or neighbored with short TEs in euchromatic regions. Collectively, our findings reveal that DNA methylation, likely acting in combination with other epigenetic modifications, may partially explain why REF6 binding is depleted in heterochromatic loci.
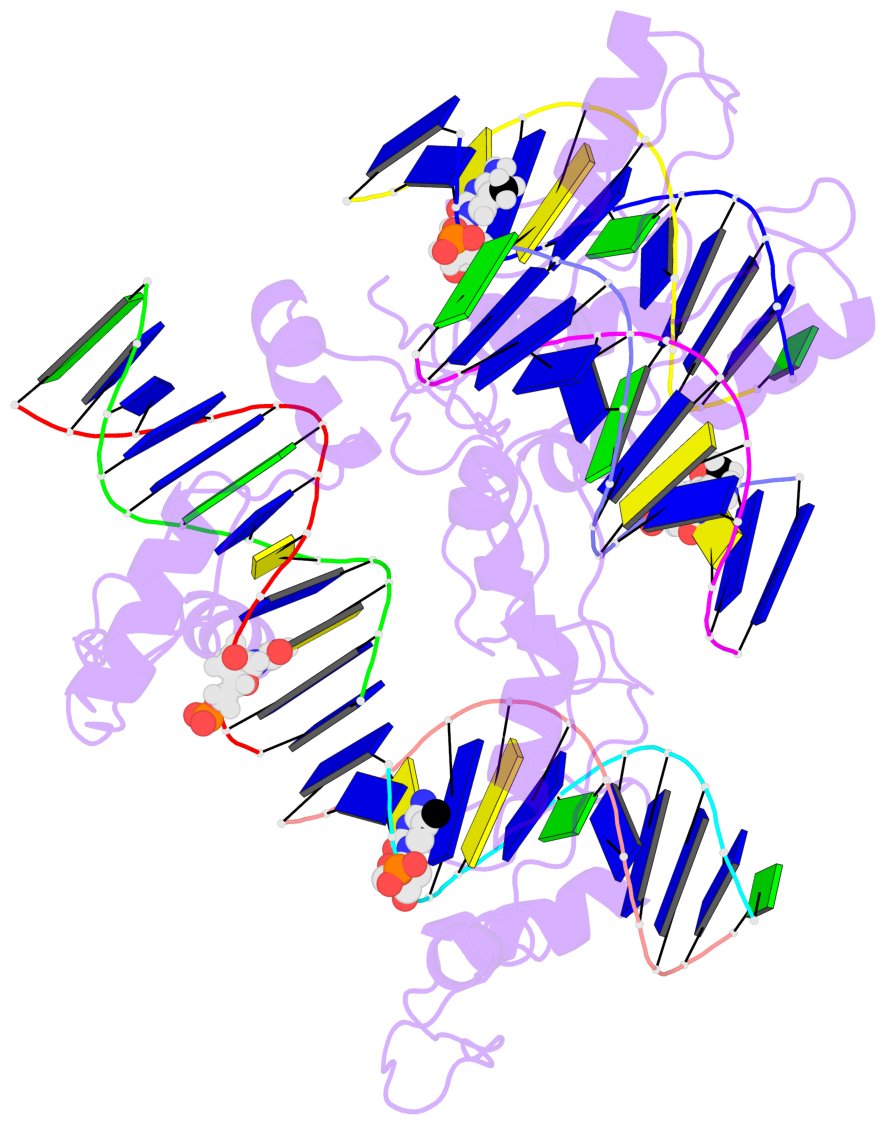
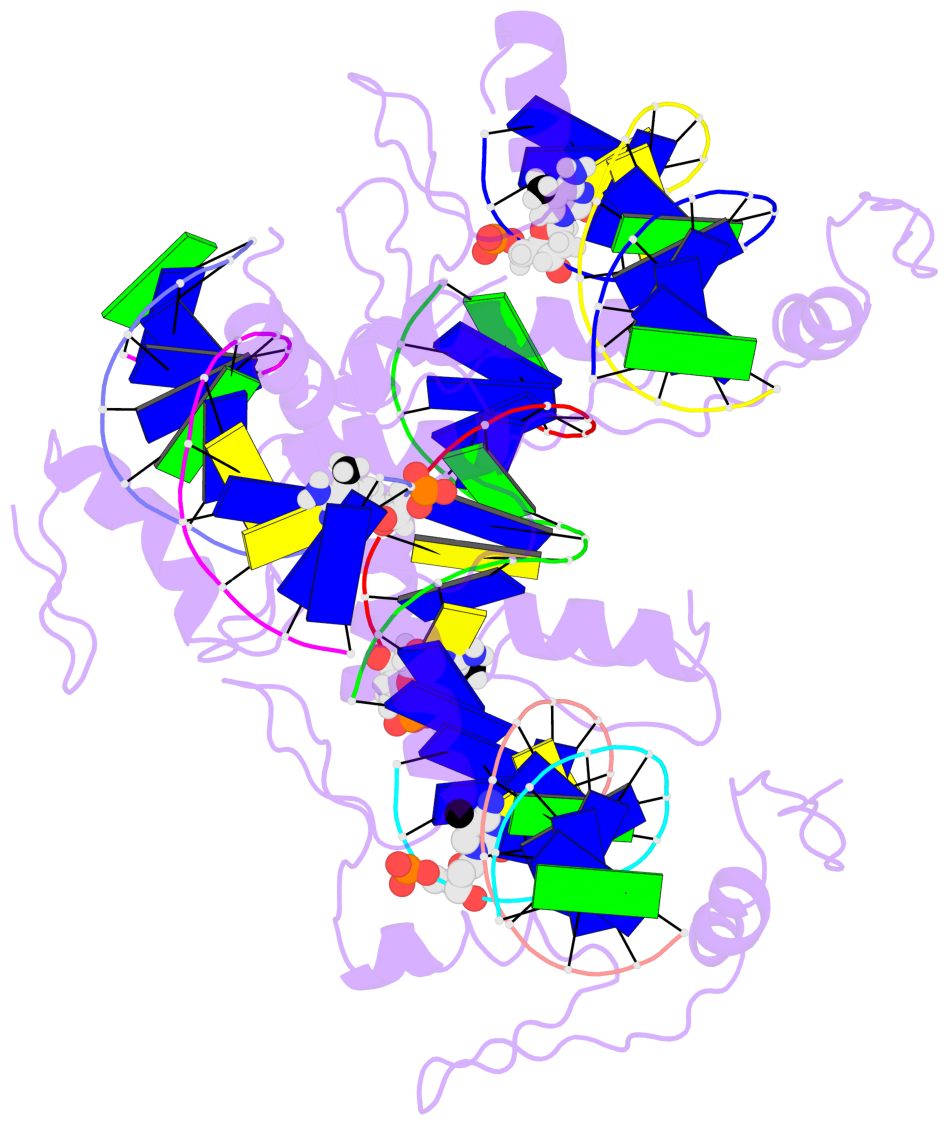
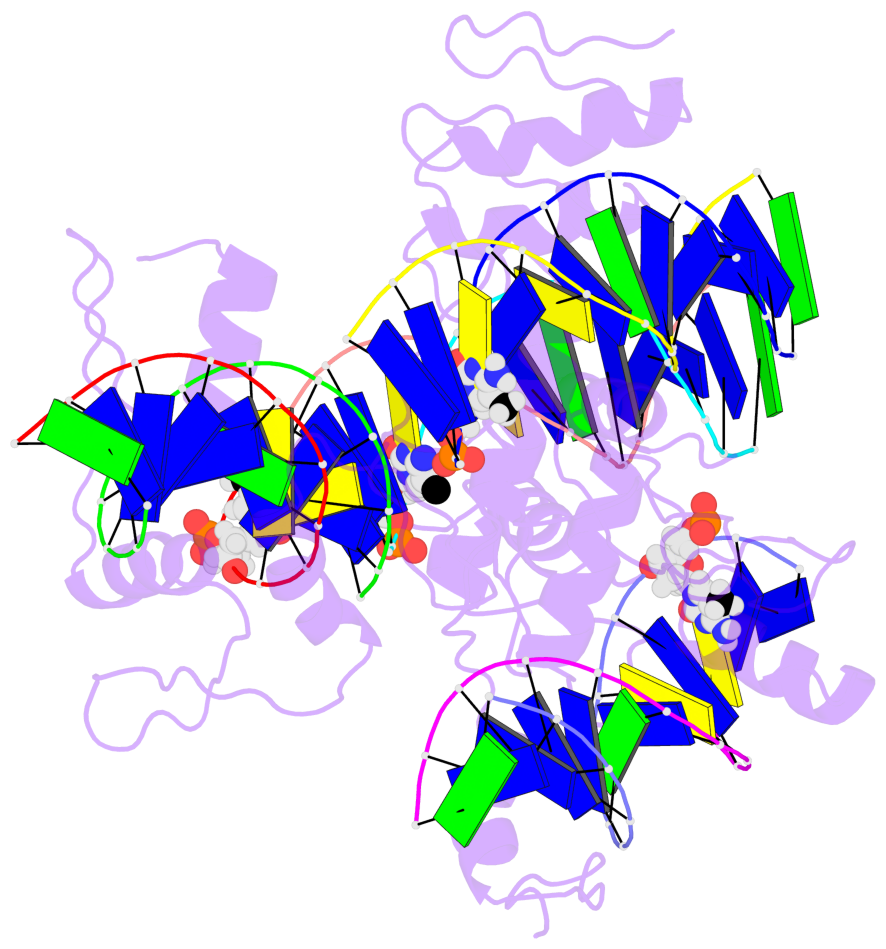
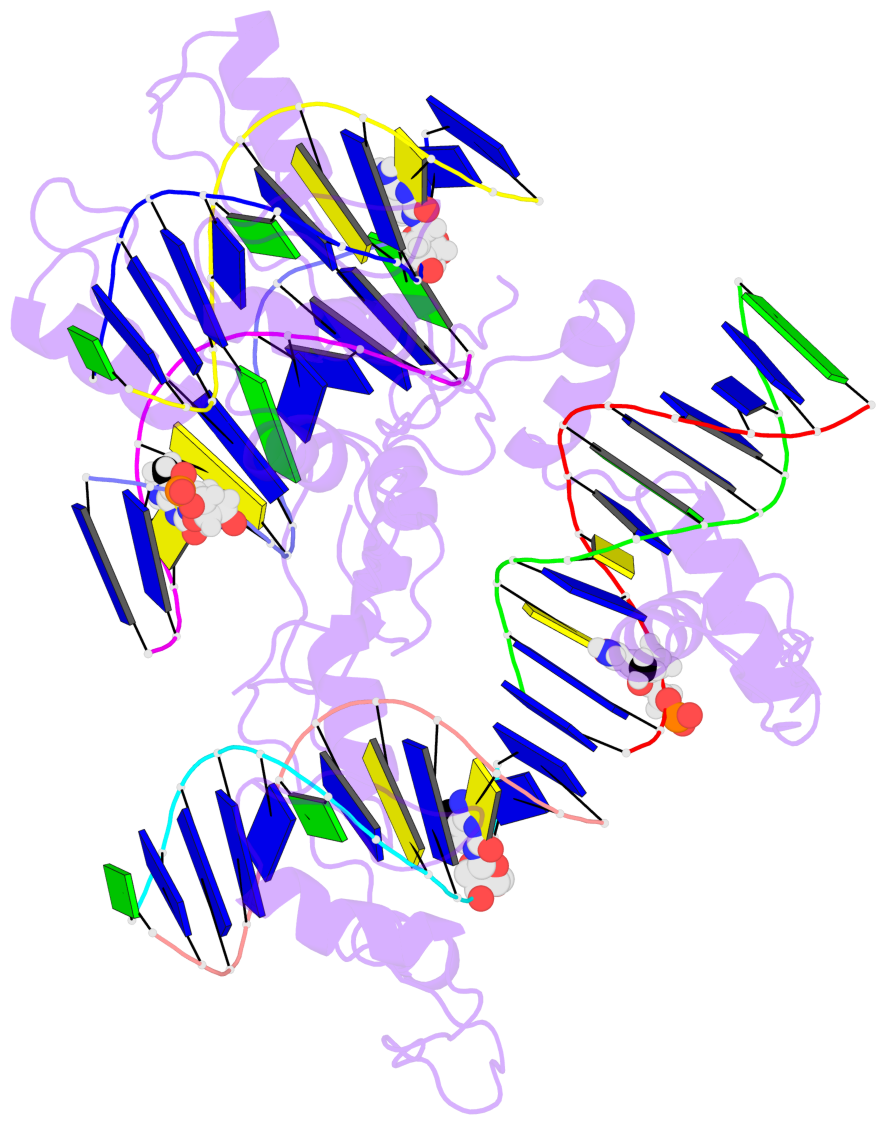
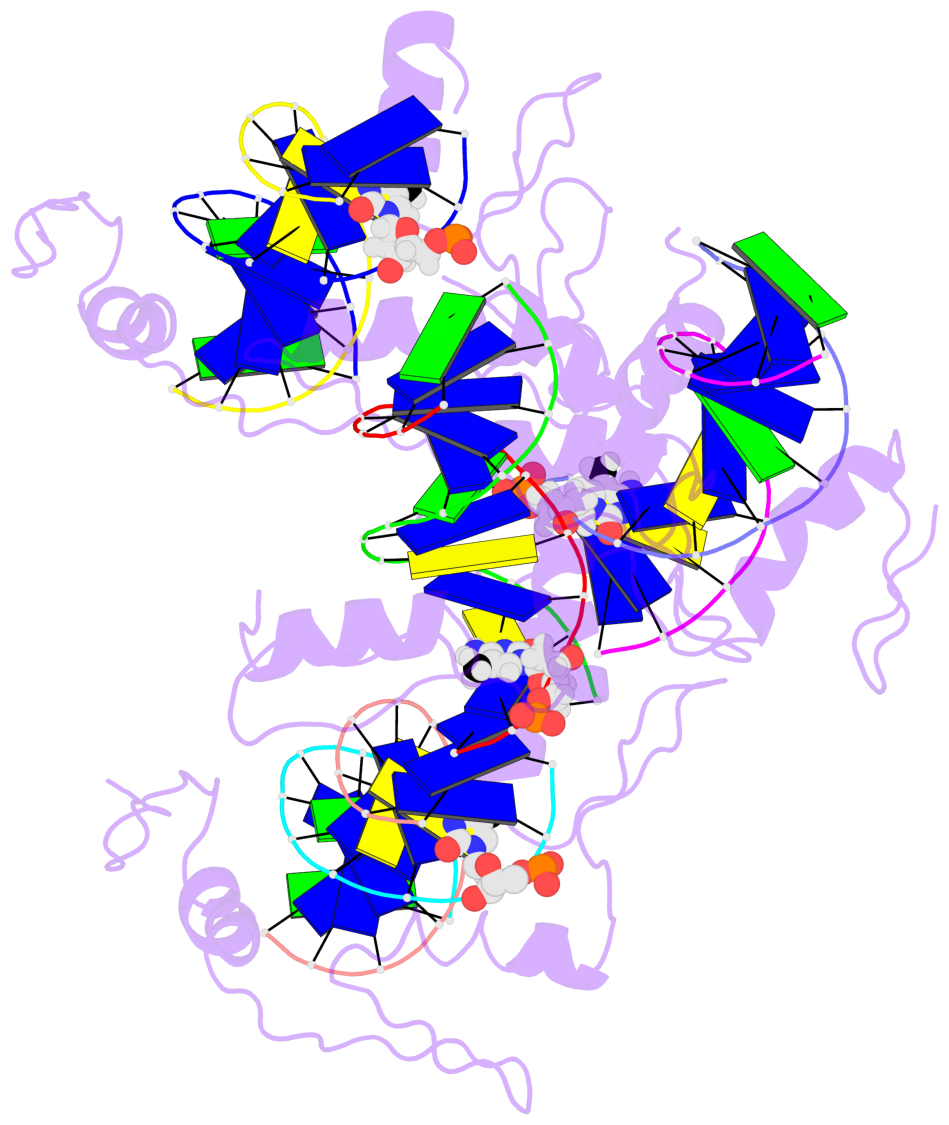
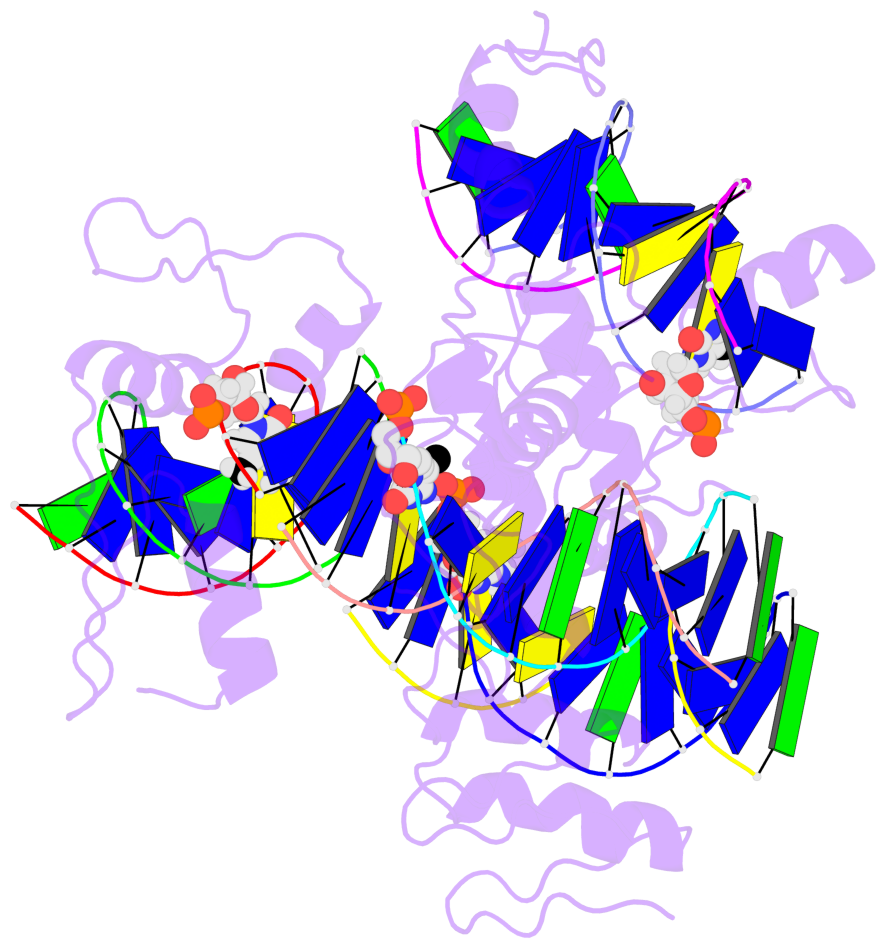
- The 5-methylcytosine group (PDB ligand '5CM') is shown in space-filling model,
with the methyl-carbon atom in black.
- Watson-Crick base pairs are represented as long rectangular blocks with the
minor-groove edge in black. Color code: A-T red, C-G yellow, G-C green, T-A blue.
- Protein is shown as cartoon in purple. DNA backbones are shown ribbon, colored code
by chain identifier.
- The block schematics were created with 3DNA-DSSR,
and images were rendered using PyMOL.
- Download the PyMOL session file corresponding to the top-left
image in the following panel.
- The contacts include paired nucleotides (mostly a G in G-C pairing), and
amino-acids within a 4.5-A distance cutoff to the base atoms of 5mC.
- The structure is oriented in the 'standard' base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
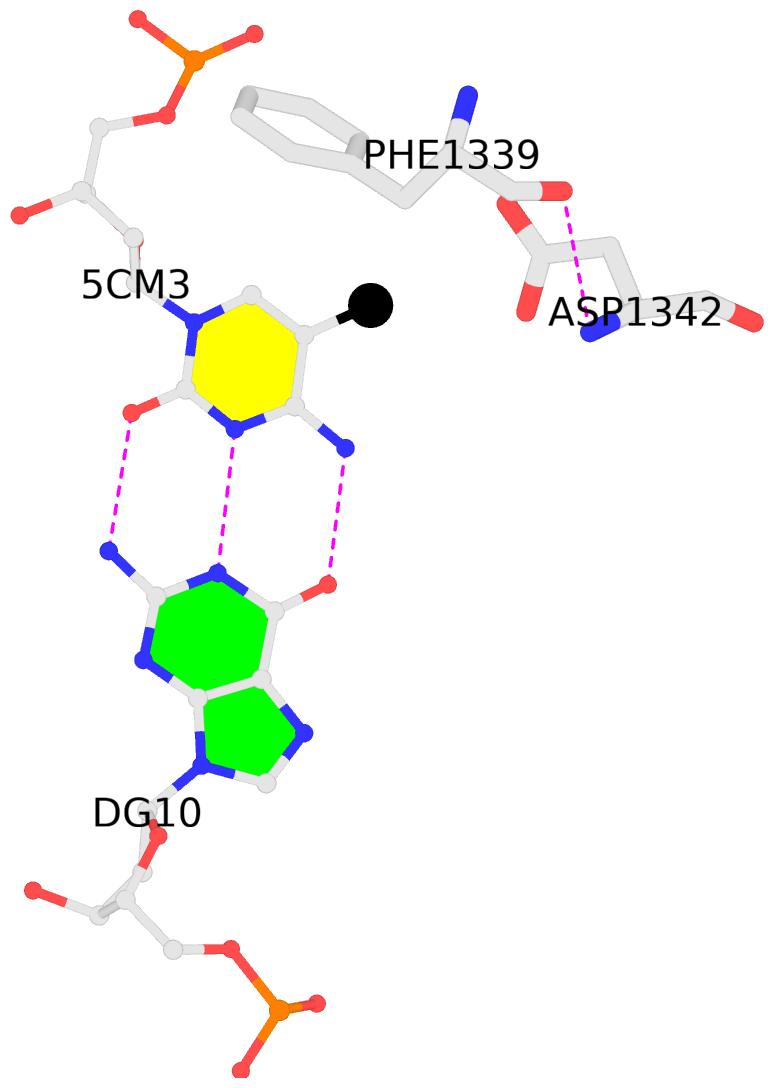 |
No. 1 D.5CM3: download PDB file
for the 5mC entry
other-contacts is-WC-paired is-in-duplex [-]:AGA/TcT
|
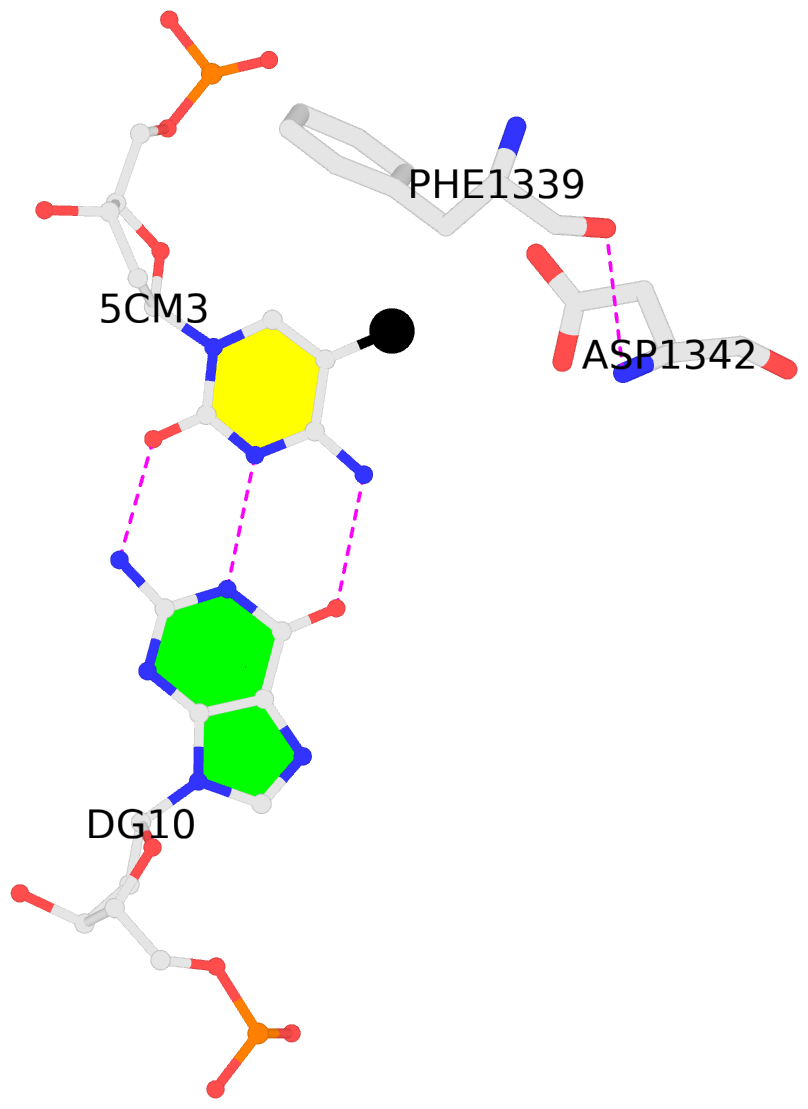 |
No. 2 F.5CM3: download PDB file
for the 5mC entry
other-contacts is-WC-paired is-in-duplex [+]:TcT/AGA
|
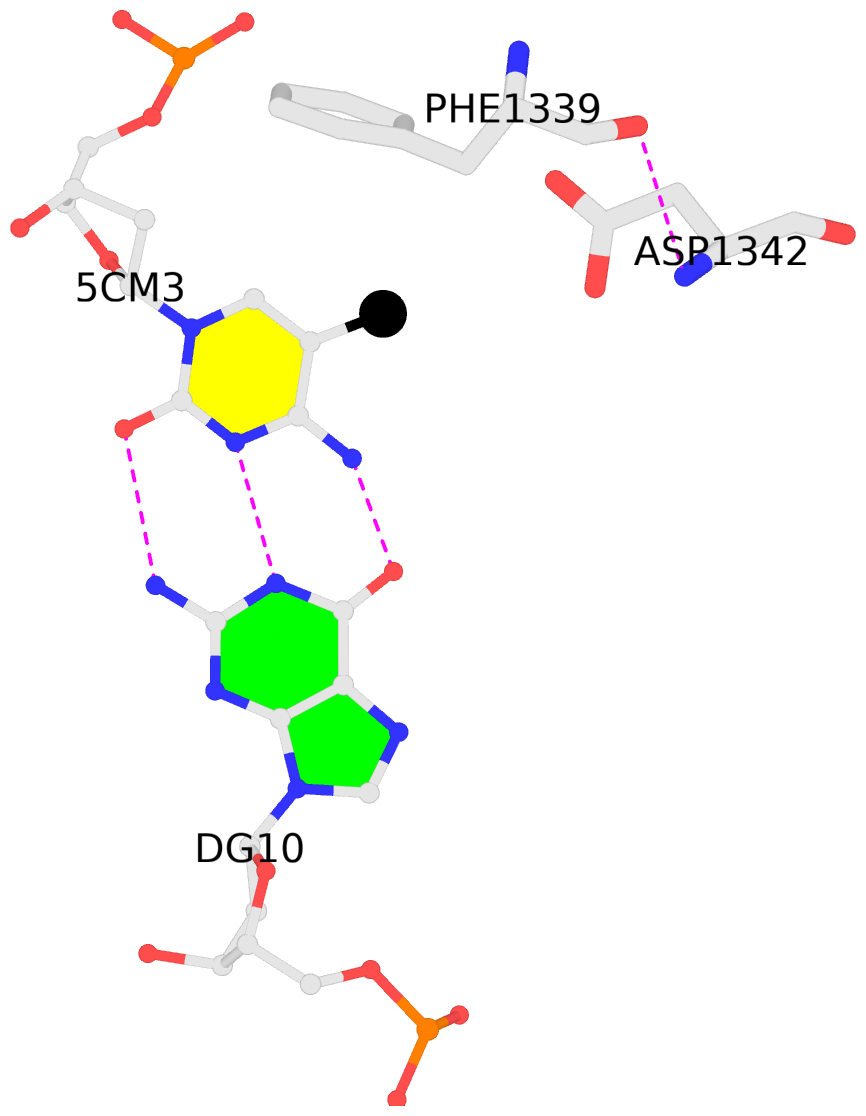 |
No. 3 I.5CM3: download PDB file
for the 5mC entry
other-contacts is-WC-paired is-in-duplex [+]:TcT/AGA
|
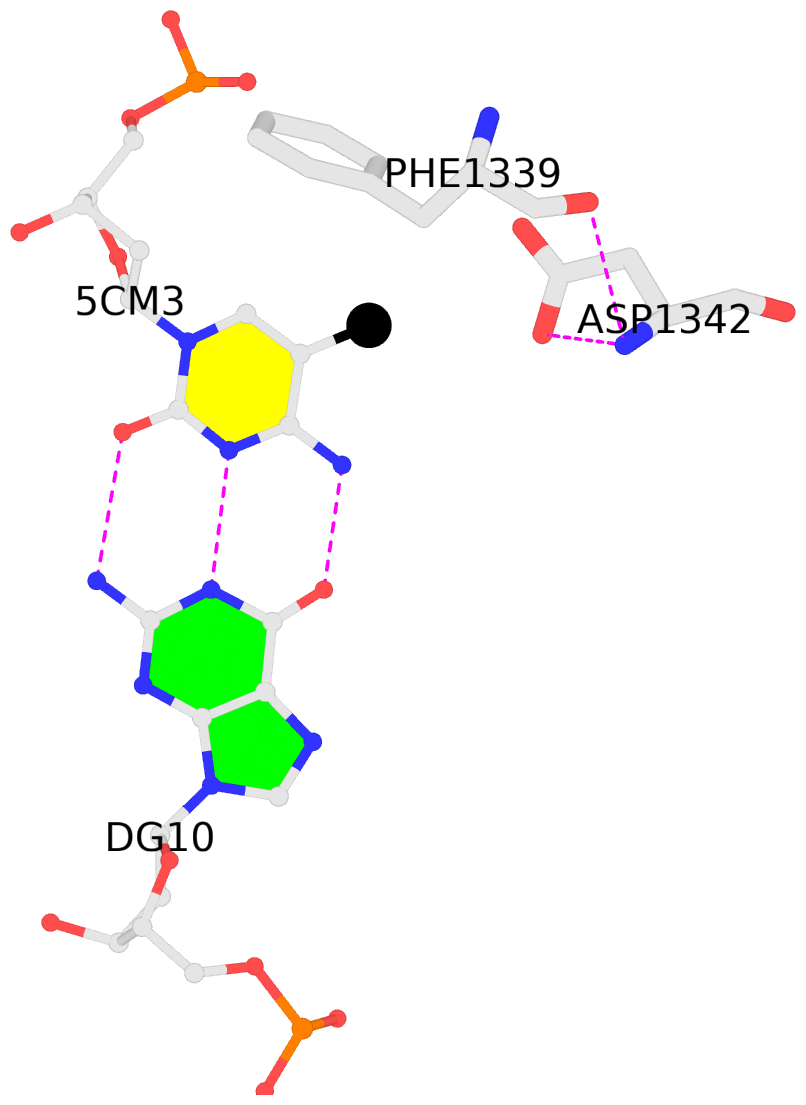 |
No. 4 L.5CM3: download PDB file
for the 5mC entry
other-contacts is-WC-paired is-in-duplex [+]:TcT/AGA
|