Last updated on 2019-09-30 by Xiang-Jun Lu <xiangjun@x3dna.org>.
The block schematics were created with DSSR and
rendered using PyMOL.
- PDB-id
- 6E93
- Class
- DNA binding protein-DNA
- Method
- X-ray (1.75 Å)
- Summary
- Crystal structure of zbtb38 c-terminal zinc fingers 6-9 in complex with methylated DNA
List of 3 5mC-amino acid contacts:
-
C.5CM24: hydrophobic-with-A.ILE1083 is-WC-paired is-in-duplex [+]:GcG/cGC
-
C.5CM27: hydrophobic-with-A.LEU1077 is-WC-paired is-in-duplex [+]:CcG/cGG
-
D.5CM9: other-contacts is-WC-paired is-in-duplex [-]:cGA/TcG
direct SNAP output · DNAproDB 2.0
- Reference
- Hudson, N.O., Whitby, F.G., Buck-Koehntop, B.A.: (2018) "Structural insights into methylated DNA recognition by the C-terminal zinc fingers of the DNA reader protein ZBTB38." J. Biol. Chem., 293, 19835-19843.
- Abstract
- Methyl-CpG-binding proteins (MBPs) are selective readers of DNA methylation that play an essential role in mediating cellular transcription processes in both normal and diseased cells. This physiological function of MBPs has generated significant interest in understanding the mechanisms by which these proteins read and interpret DNA methylation signals. Zinc finger and BTB domain-containing 38 (ZBTB38) represents one member of the zinc finger (ZF) family of MBPs. We recently demonstrated that the C-terminal ZFs of ZBTB38 exhibit methyl-selective DNA binding within the ((A/G)TmCG(G/A)(mC/T)(G/A)) context both in vitro and within cells. Here we report the crystal structure of the first four C-terminal ZBTB38 ZFs (ZFs 6-9) in complex with the previously identified methylated consensus sequence at 1.75 Å resolution. From the structure, methyl-selective binding is preferentially localized at the 5' mCpG site of the bound DNA, which is facilitated through a series of base-specific interactions from residues within the α-helices of ZF7 and ZF8. ZF6 and ZF9 primarily stabilize ZF7 and ZF8 to facilitate the core base-specific interactions. Further structural and biochemical analyses, including solution NMR spectroscopy and electrophoretic mobility gel shift assays, revealed that the C-terminal ZFs of ZBTB38 utilize an alternative mode of mCpG recognition from the ZF MBPs structurally evaluated to date. Combined, these findings provide insight into the mechanism by which this ZF domain of ZBTB38 selectively recognizes methylated CpG sites and expands our understanding of how ZF-containing proteins can interpret this essential epigenetic mark.
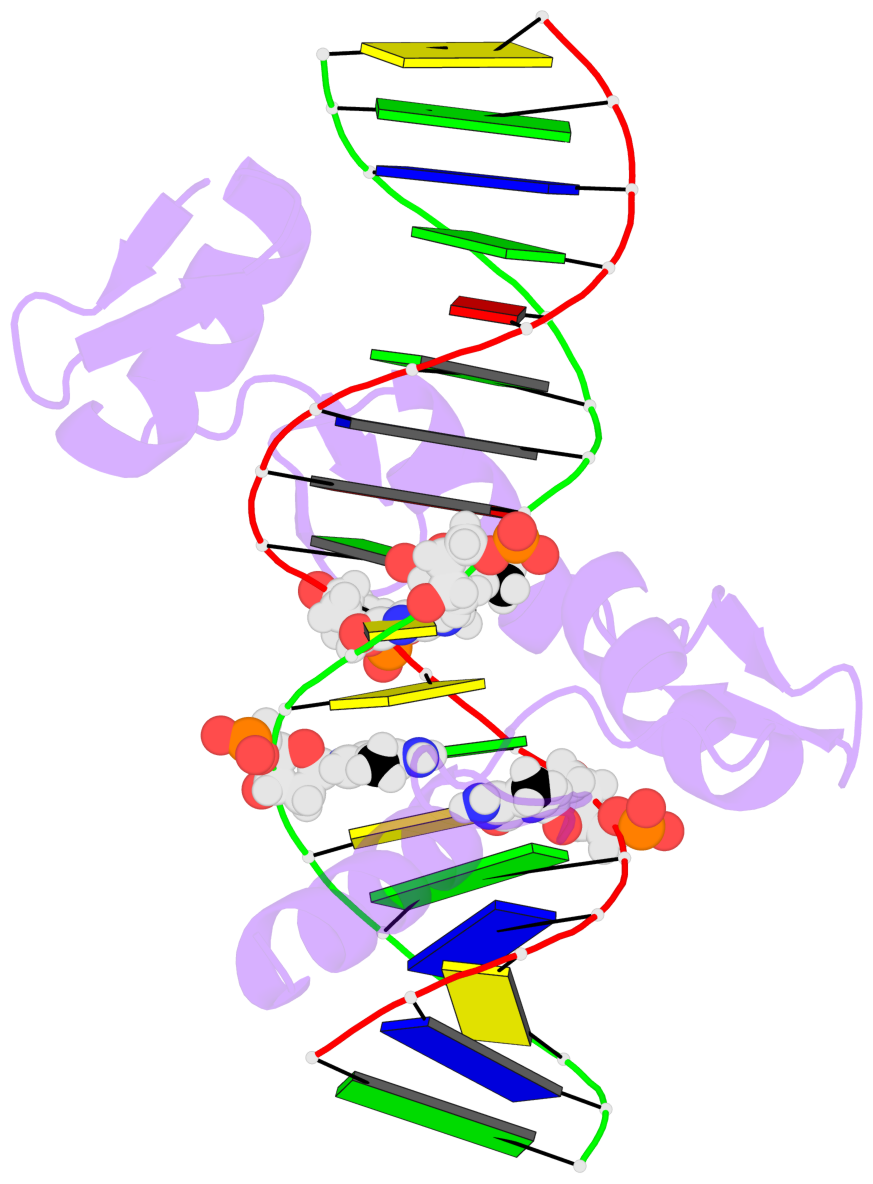
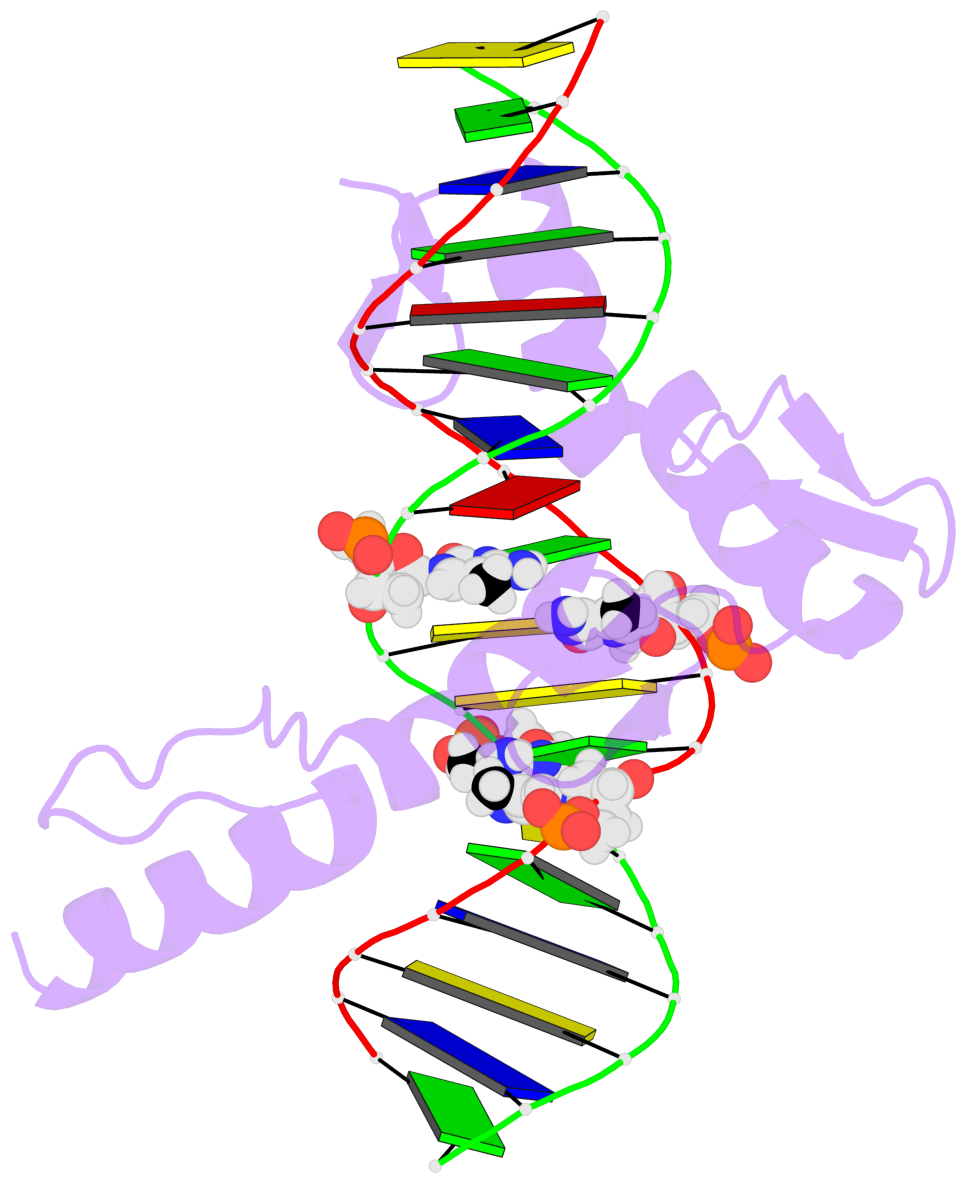
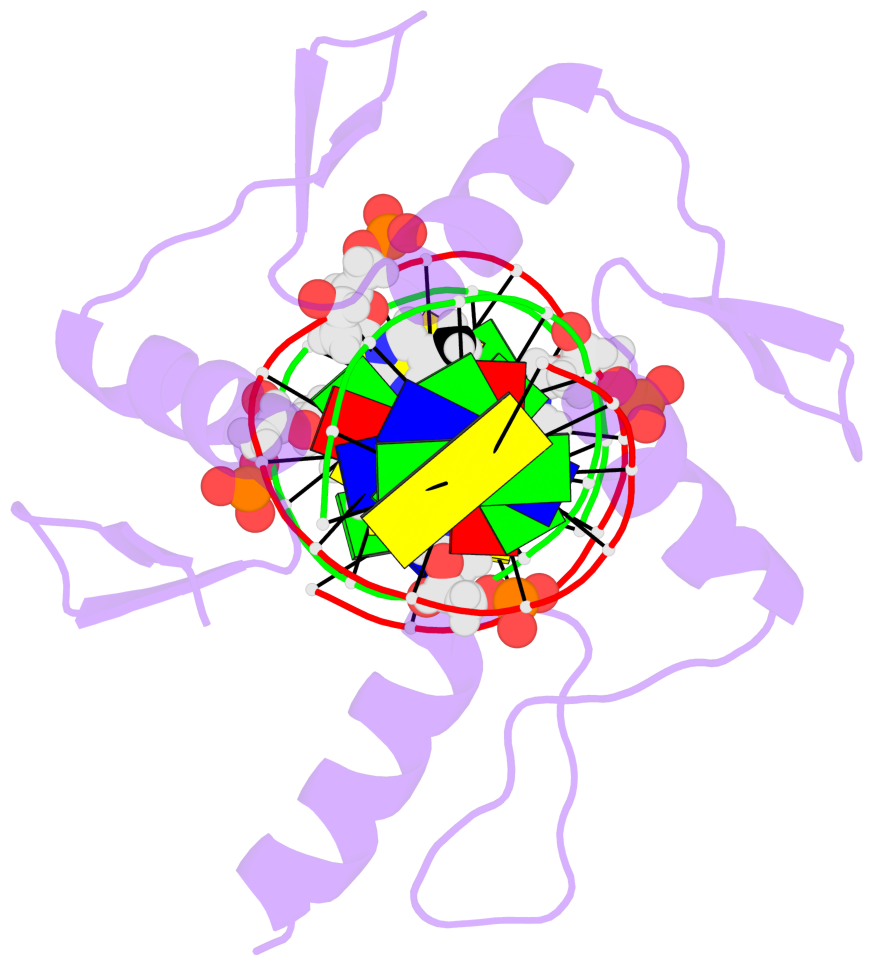
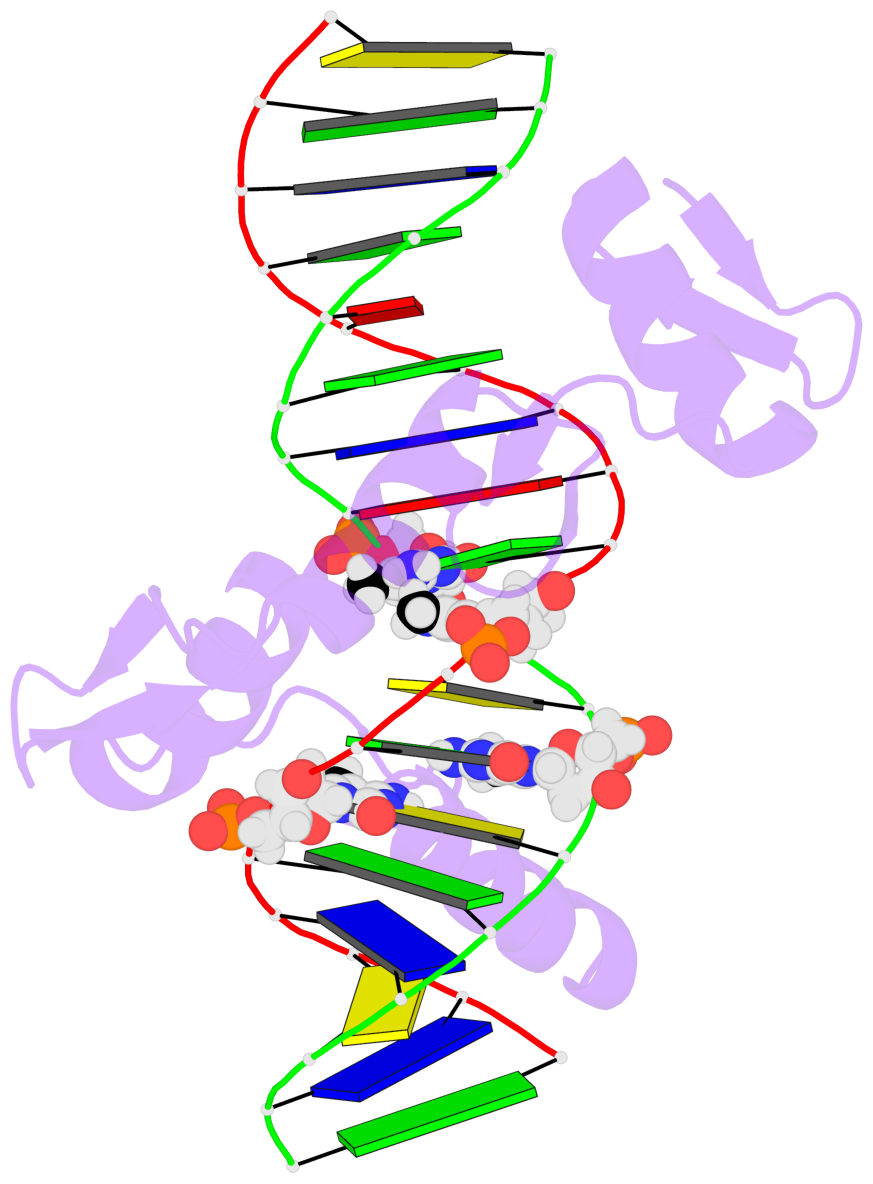
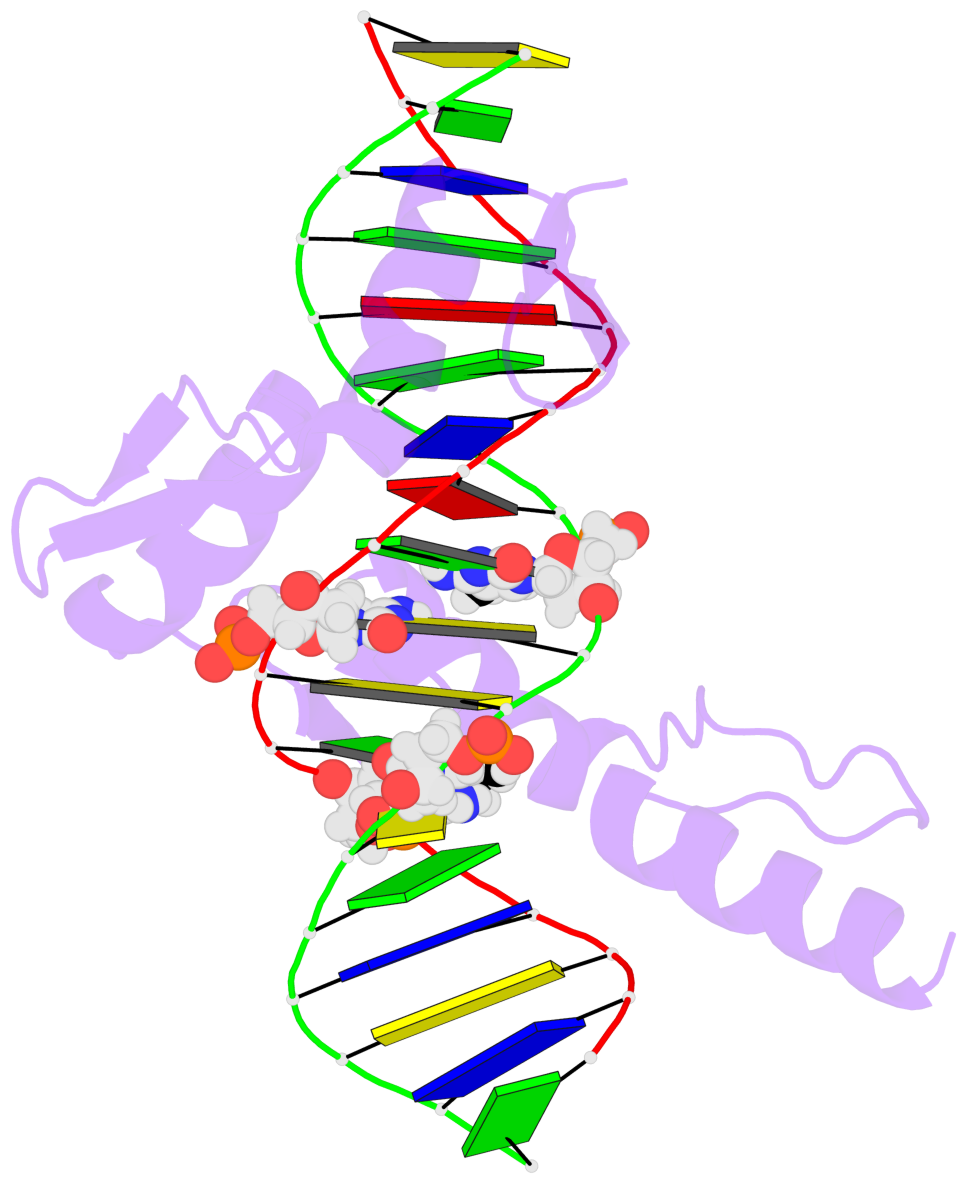
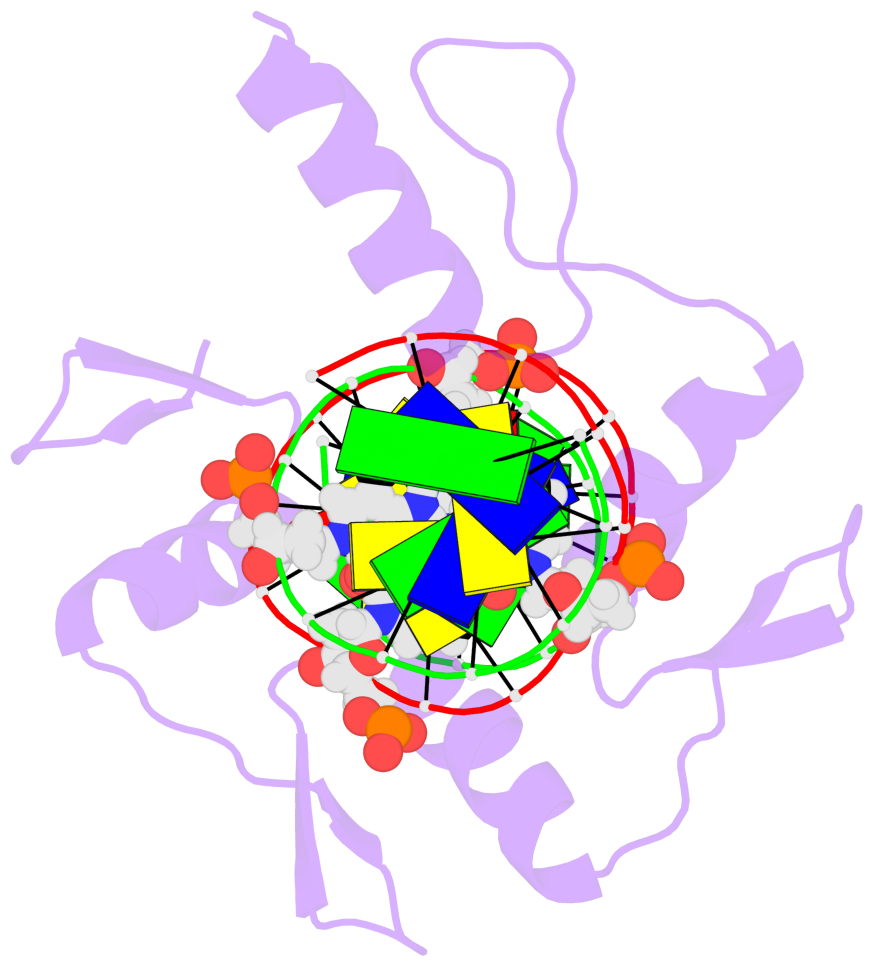
- The 5-methylcytosine group (PDB ligand '5CM') is shown in space-filling model,
with the methyl-carbon atom in black.
- Watson-Crick base pairs are represented as long rectangular blocks with the
minor-groove edge in black. Color code: A-T red, C-G yellow, G-C green, T-A blue.
- Protein is shown as cartoon in purple. DNA backbones are shown ribbon, colored code
by chain identifier.
- The block schematics were created with 3DNA-DSSR,
and images were rendered using PyMOL.
- Download the PyMOL session file corresponding to the top-left
image in the following panel.
- The contacts include paired nucleotides (mostly a G in G-C pairing), and
amino-acids within a 4.5-A distance cutoff to the base atoms of 5mC.
- The structure is oriented in the 'standard' base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
 |
No. 1 C.5CM24: download PDB file
for the 5mC entry
hydrophobic-with-A.ILE1083 is-WC-paired is-in-duplex [+]:GcG/cGC
|
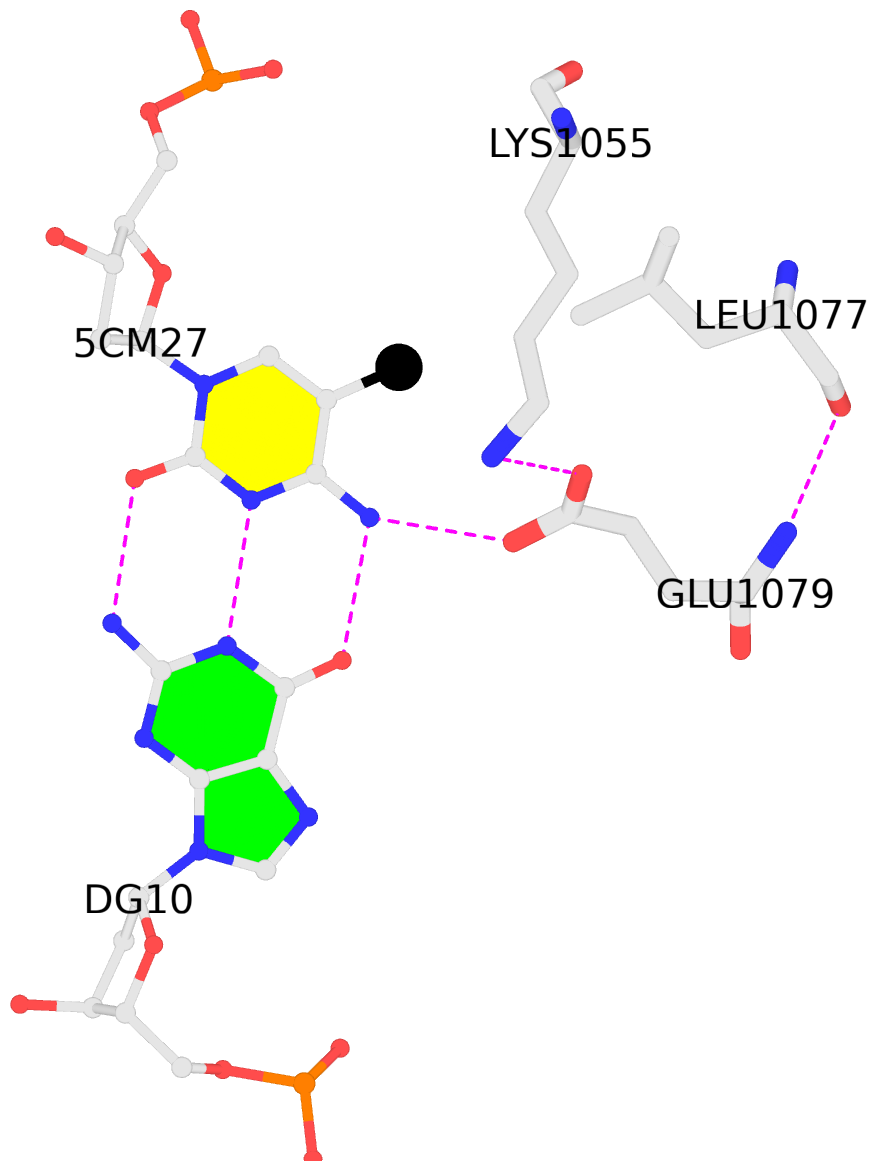 |
No. 2 C.5CM27: download PDB file
for the 5mC entry
hydrophobic-with-A.LEU1077 is-WC-paired is-in-duplex [+]:CcG/cGG
|
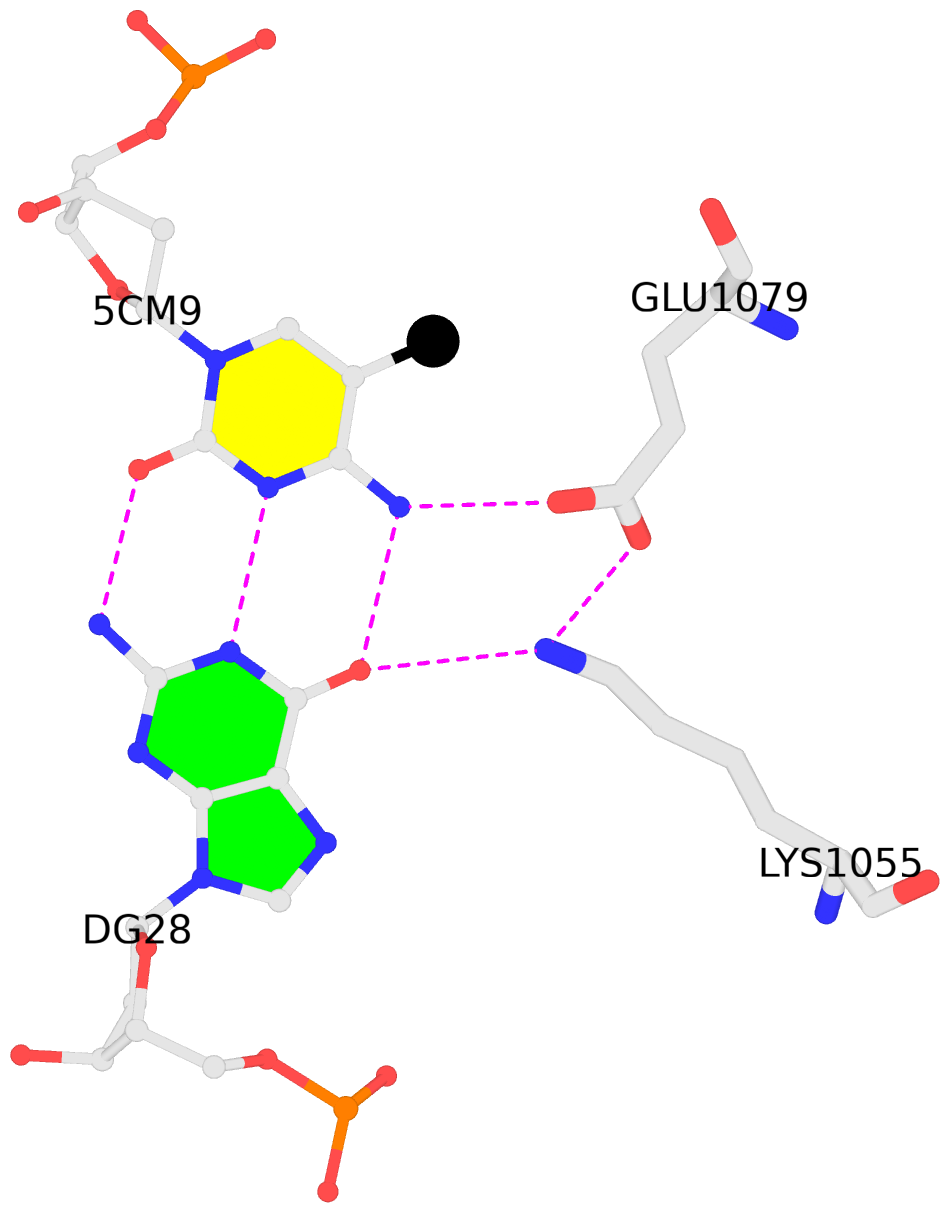 |
No. 3 D.5CM9: download PDB file
for the 5mC entry
other-contacts is-WC-paired is-in-duplex [-]:cGA/TcG
|