Last updated on 2019-09-30 by Xiang-Jun Lu <xiangjun@x3dna.org>.
The block schematics were created with DSSR and
rendered using PyMOL.
- PDB-id
- 6C1T
- Class
- DNA binding protein-DNA
- Method
- X-ray (1.84 Å)
- Summary
- Mbd2 in complex with a partially methylated DNA
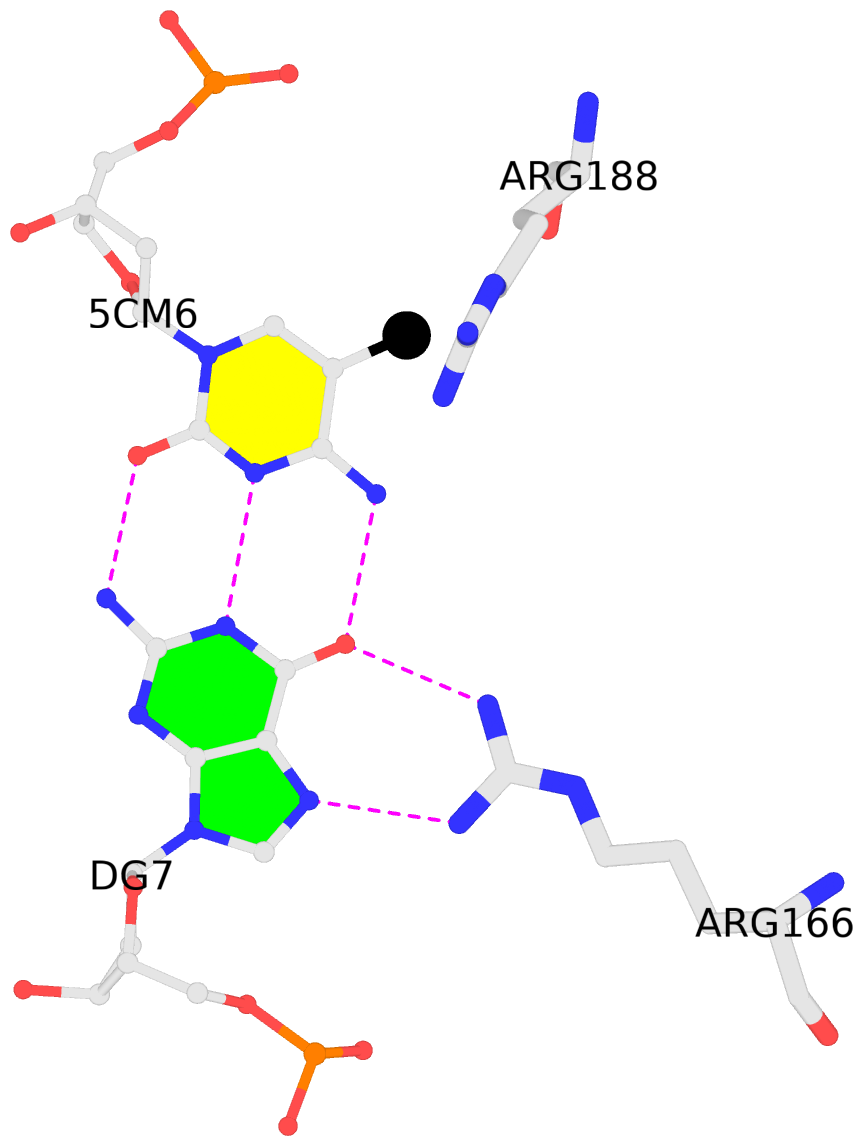
List of 1 5mC-amino acid contact:
-
B.5CM6: other-contacts is-WC-paired is-in-duplex [+]:AcA/TGT
direct SNAP output · DNAproDB 2.0
- Reference
- Liu, K., Xu, C., Lei, M., Yang, A., Loppnau, P., Hughes, T.R., Min, J.: (2018) "Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA." J. Biol. Chem., 293, 7344-7354.
- Abstract
- Cytosine methylation is a well-characterized epigenetic mark and occurs at both CG and non-CG sites in DNA. Both methylated CG (mCG)- and mCH (H = A, C, or T)-containing DNAs, especially mCAC-containing DNAs, are recognized by methyl-CpG-binding protein 2 (MeCP2) to regulate gene expression in neuron development. However, the molecular mechanism involved in the binding of methyl-CpG-binding domain (MBD) of MeCP2 to these different DNA motifs is unclear. Here, we systematically characterized the DNA-binding selectivities of the MBD domains in MeCP2 and MBD1-4 with isothermal titration calorimetry-based binding assays, mutagenesis studies, and X-ray crystallography. We found that the MBD domains of MeCP2 and MBD1-4 bind mCG-containing DNAs independently of the sequence identity outside the mCG dinucleotide. Moreover, some MBD domains bound to both methylated and unmethylated CA dinucleotide-containing DNAs, with a preference for the CAC sequence motif. We also found that the MBD domains bind to mCA or nonmethylated CA DNA by recognizing the complementary TG dinucleotide, which is consistent with an overlooked ligand of MeCP2, i.e. the matrix/scaffold attachment regions (MARs/SARs) with a consensus sequence of 5'-GGTGT-3' that was identified in early 1990s. Our results also explain why MeCP2 exhibits similar binding affinity to both mCA- and hmCA-containing dsDNAs. In summary, our results suggest that in addition to mCG sites, unmethylated CA or TG sites also serve as DNA-binding sites for MeCP2 and other MBD-containing proteins. This discovery expands the genome-wide activity of MBD-containing proteins in gene regulation.
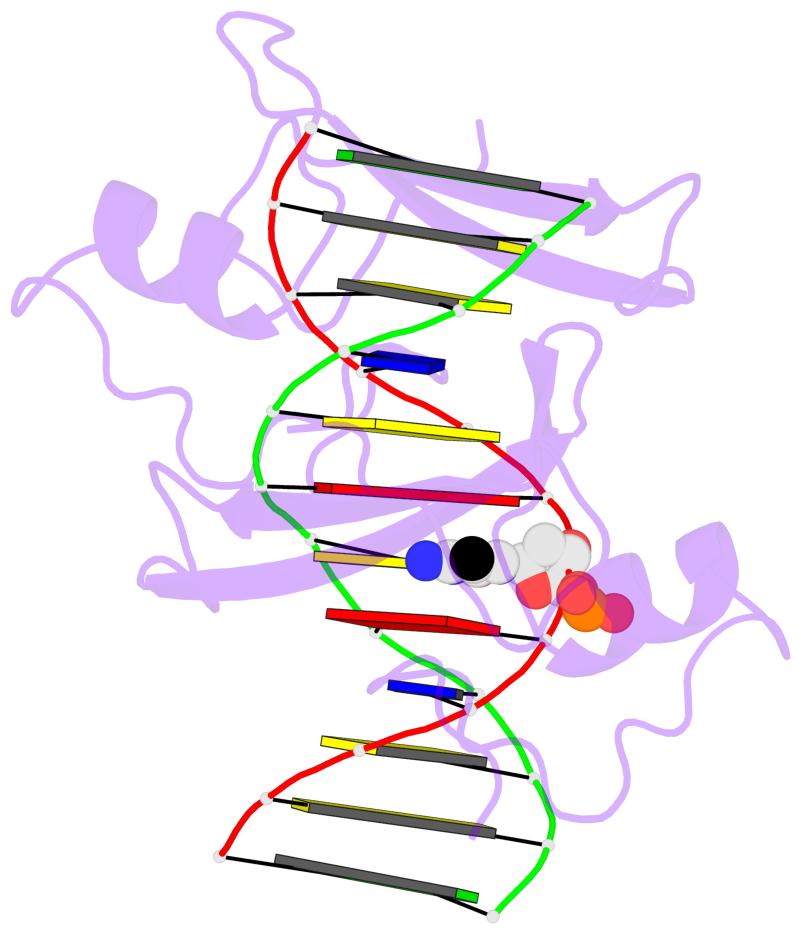
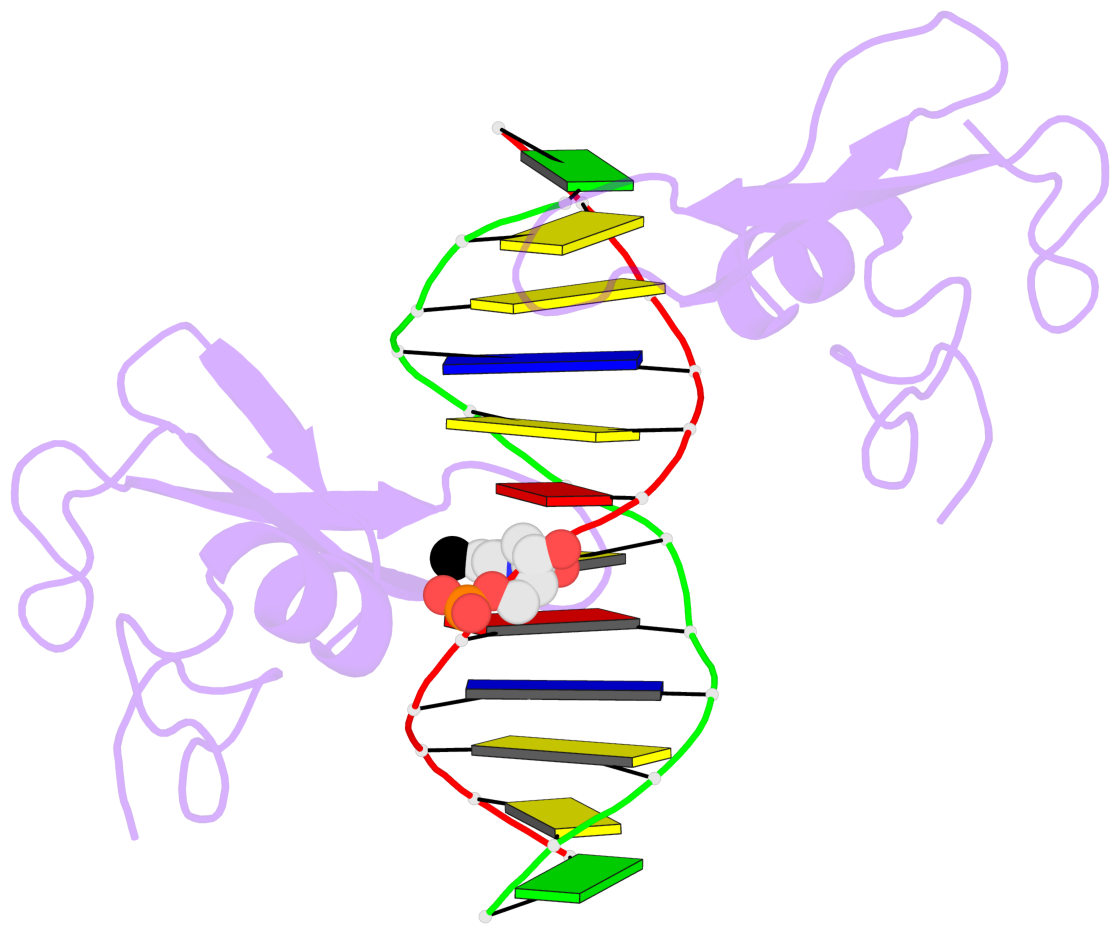
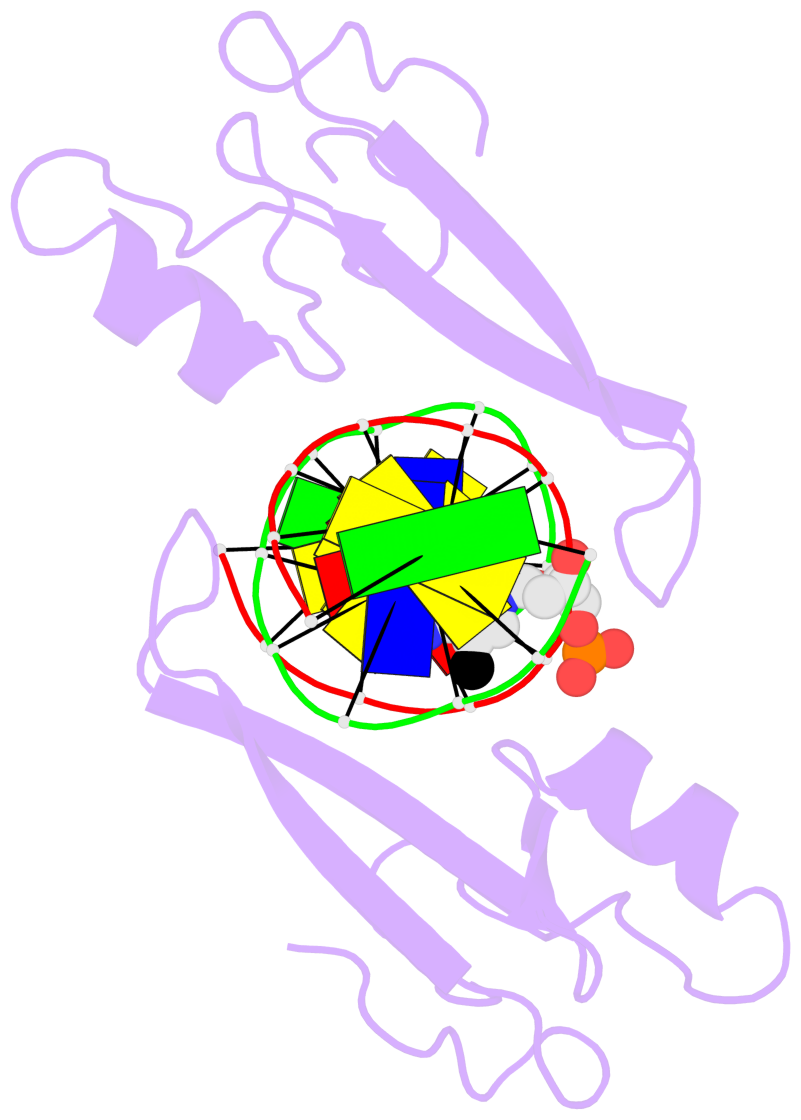
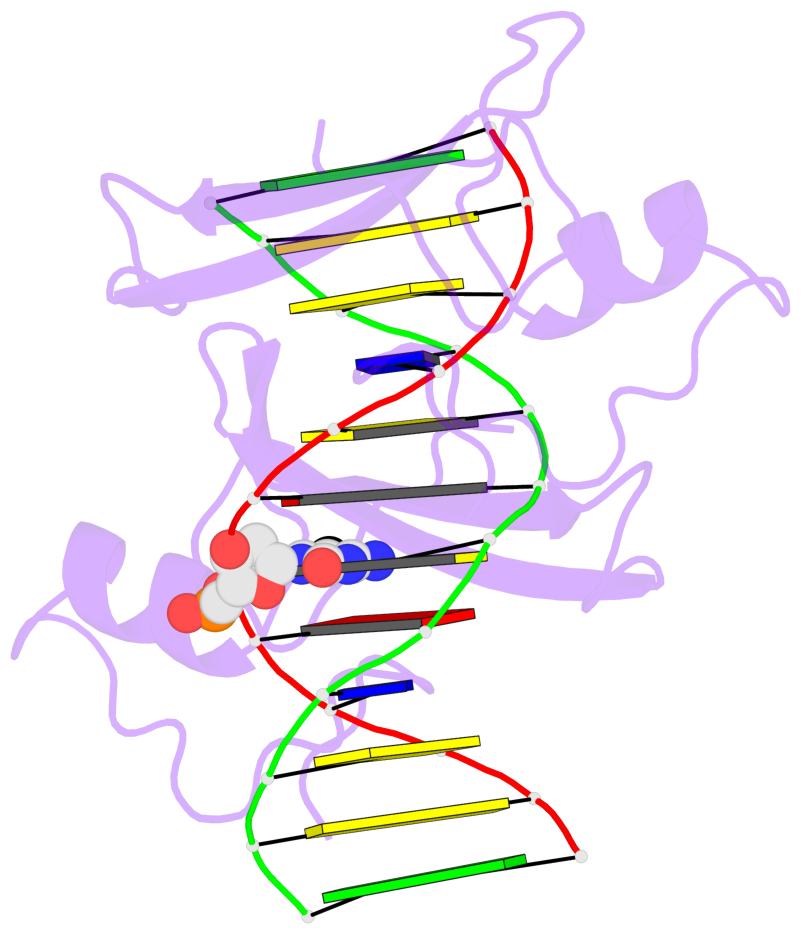
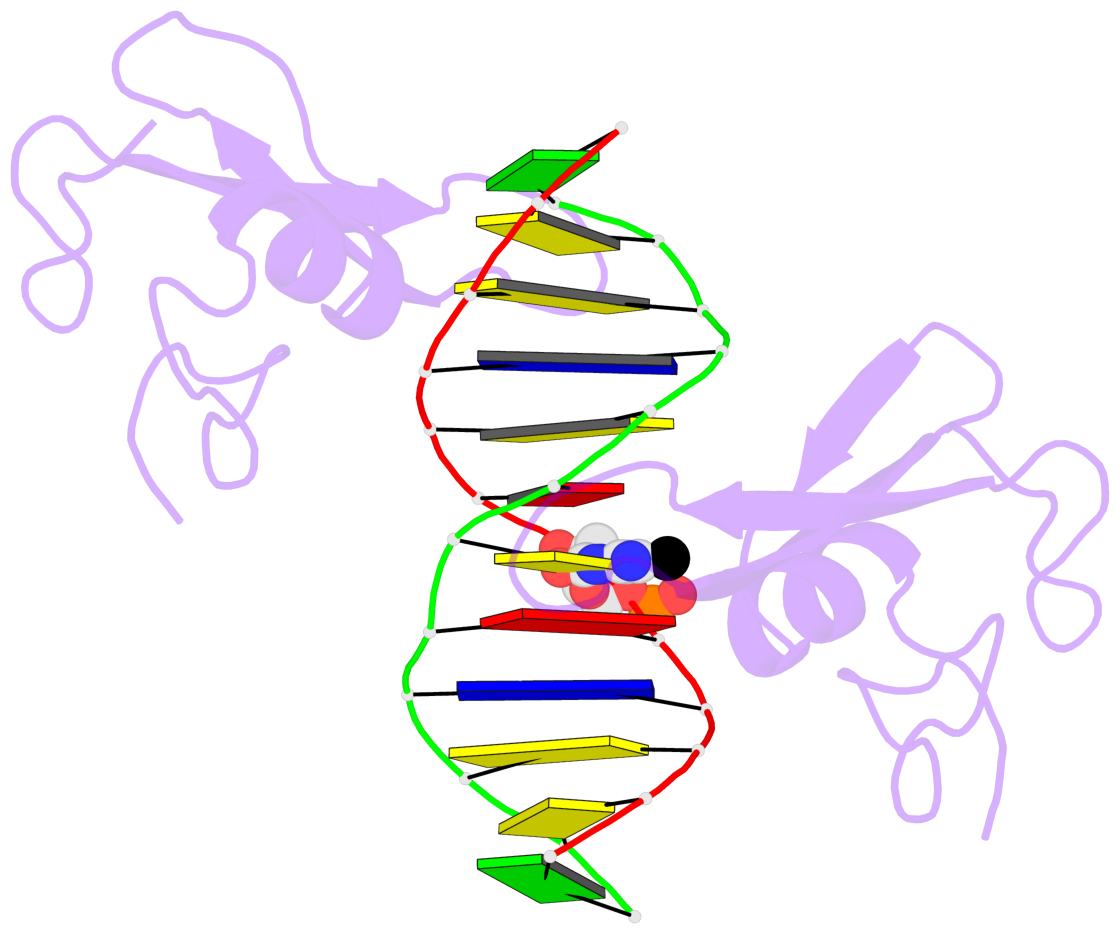
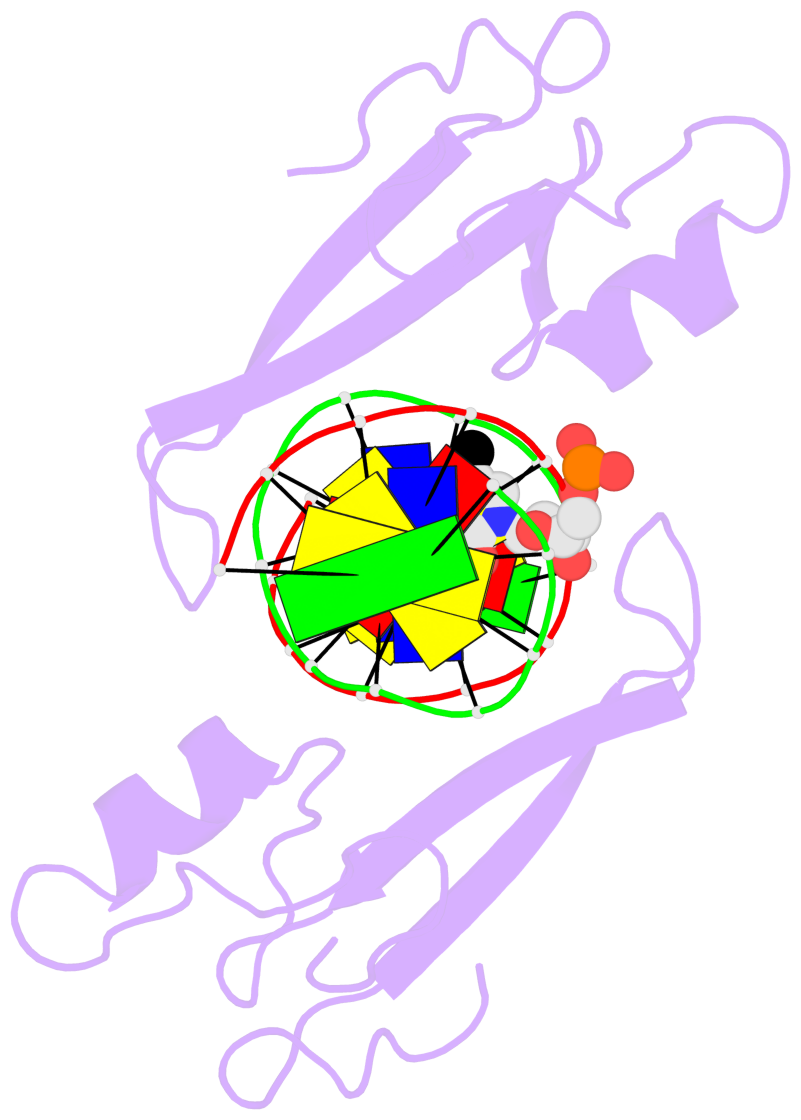
- The 5-methylcytosine group (PDB ligand '5CM') is shown in space-filling model,
with the methyl-carbon atom in black.
- Watson-Crick base pairs are represented as long rectangular blocks with the
minor-groove edge in black. Color code: A-T red, C-G yellow, G-C green, T-A blue.
- Protein is shown as cartoon in purple. DNA backbones are shown ribbon, colored code
by chain identifier.
- The block schematics were created with 3DNA-DSSR,
and images were rendered using PyMOL.
- Download the PyMOL session file corresponding to the top-left
image in the following panel.
- The contacts include paired nucleotides (mostly a G in G-C pairing), and
amino-acids within a 4.5-A distance cutoff to the base atoms of 5mC.
- The structure is oriented in the 'standard' base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
 |
No. 1 B.5CM6: download PDB file
for the 5mC entry
other-contacts is-WC-paired is-in-duplex [+]:AcA/TGT
|