Last updated on 2019-09-30 by Xiang-Jun Lu <xiangjun@x3dna.org>.
The block schematics were created with DSSR and
rendered using PyMOL.
- PDB-id
- 6A5N
- Class
- gene regulation-DNA
- Method
- X-ray (2.4 Å)
- Summary
- Crystal structure of arabidopsis thaliana suvh6 in complex with methylated DNA
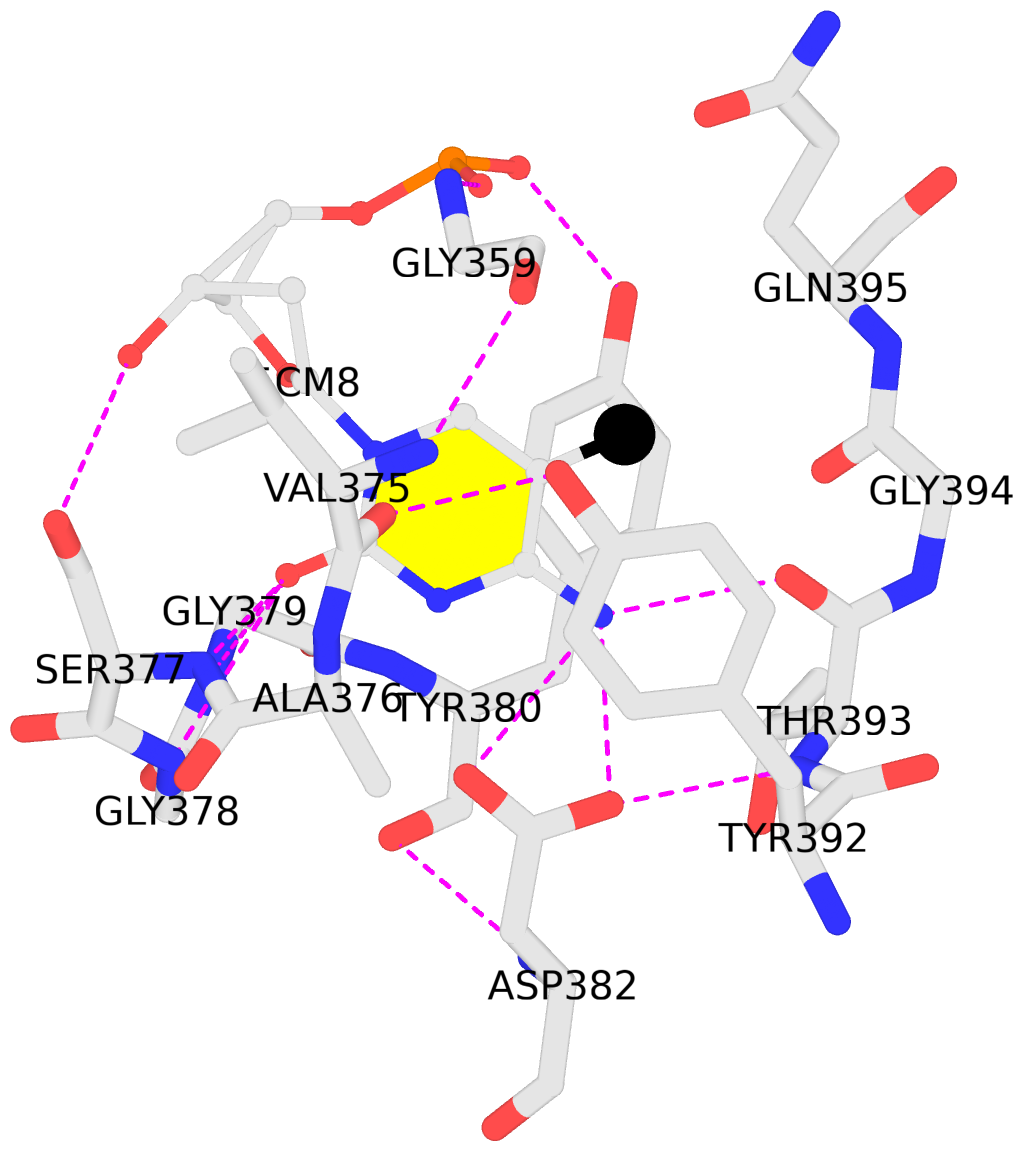
List of 1 5mC-amino acid contact:
-
C.5CM8: stacking-with-A.TYR380 stacking-with-A.TYR392 not-WC-paired not-in-duplex
direct SNAP output · DNAproDB 2.0
- Reference
- Li, X., Harris, C.J., Zhong, Z., Chen, W., Liu, R., Jia, B., Wang, Z., Li, S., Jacobsen, S.E., Du, J.: (2018) "Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation." Proc. Natl. Acad. Sci. U.S.A., 115, E8793-E8802.
- Abstract
- DNA methylation functions in gene silencing and the maintenance of genome integrity. In plants, non-CG DNA methylation is linked through a self-reinforcing loop with histone 3 lysine 9 dimethylation (H3K9me2). The plant-specific SUPPRESSOR OF VARIEGATION 3-9 HOMOLOG (SUVH) family H3K9 methyltransferases (MTases) bind to DNA methylation marks and catalyze H3K9 methylation. Here, we analyzed the structure and function of Arabidopsis thaliana SUVH6 to understand how this class of enzyme maintains methylation patterns in the genome. We reveal that SUVH6 has a distinct 5-methyl-dC (5mC) base-flipping mechanism involving a thumb loop element. Autoinhibition of H3 substrate entry is regulated by a SET domain loop, and a conformational transition in the post-SET domain upon cofactor binding may control catalysis. In vitro DNA binding and in vivo ChIP-seq data reveal that the different SUVH family H3K9 MTases have distinct DNA binding preferences, targeting H3K9 methylation to sites with different methylated DNA sequences, explaining the context biased non-CG DNA methylation in plants.
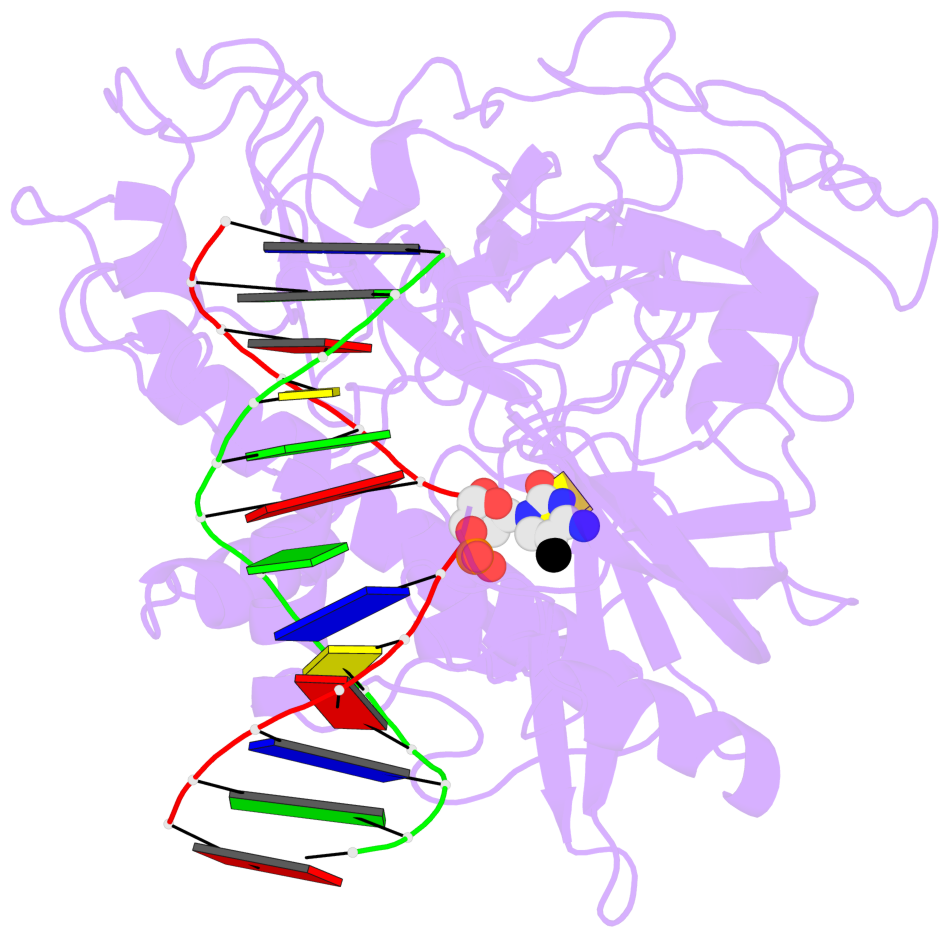
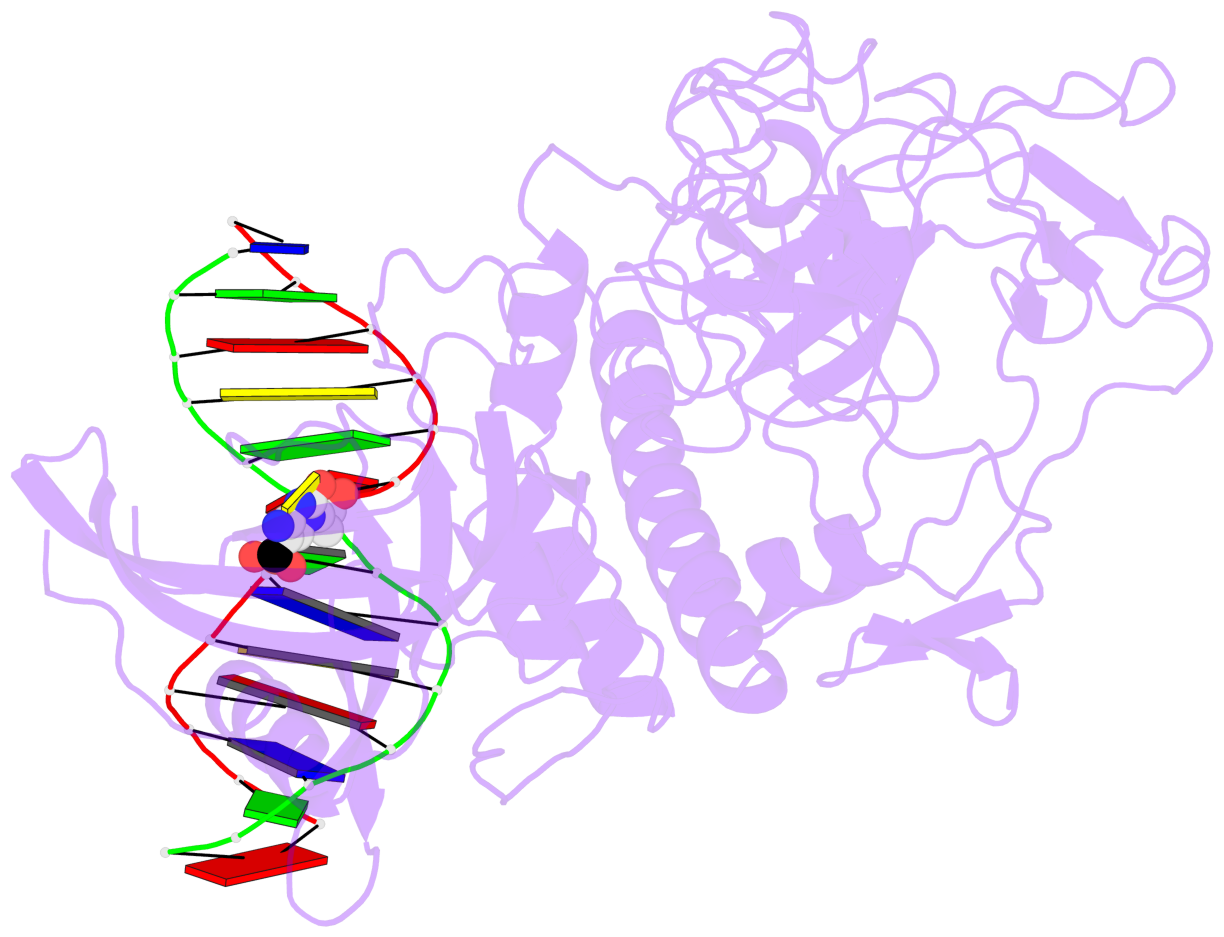
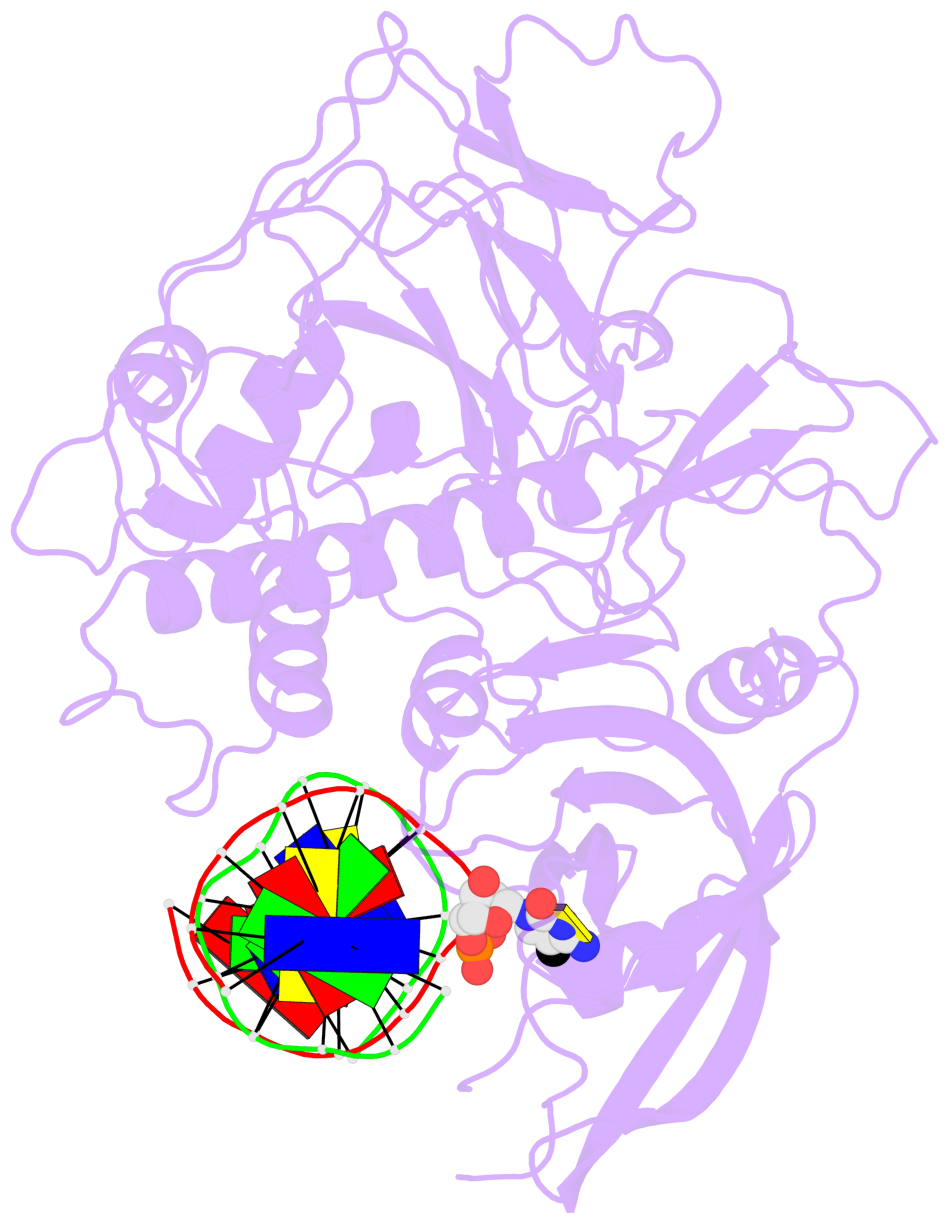
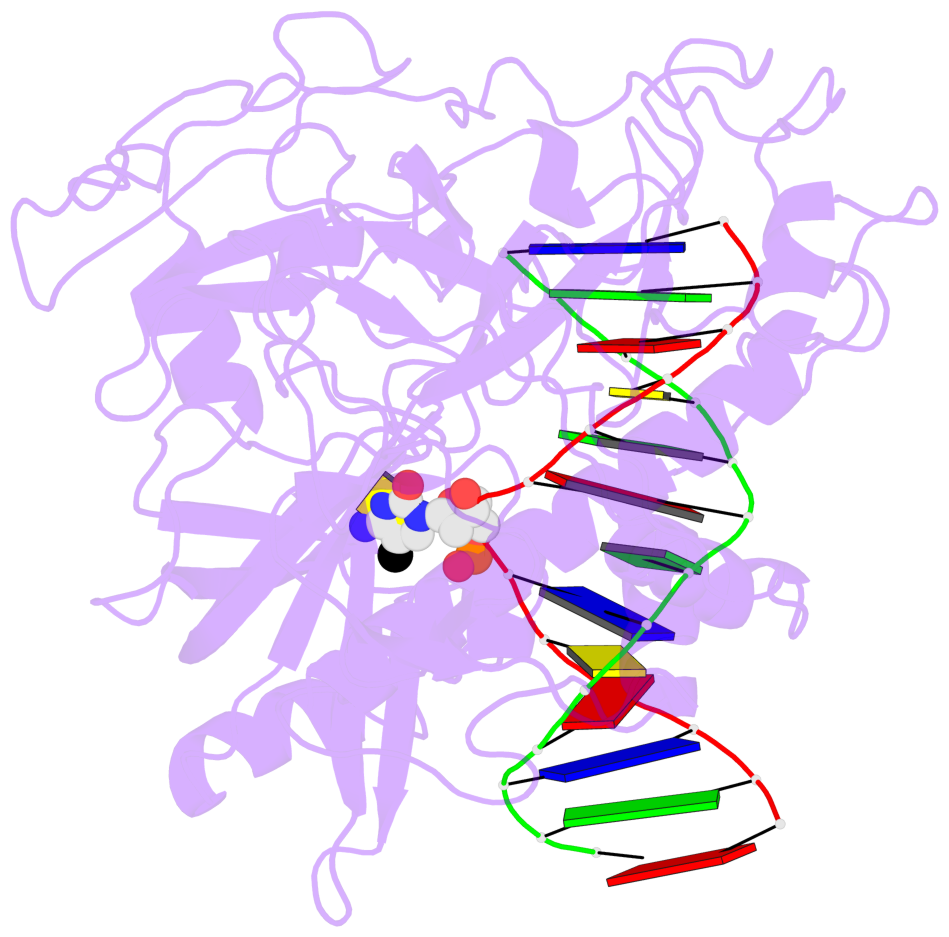
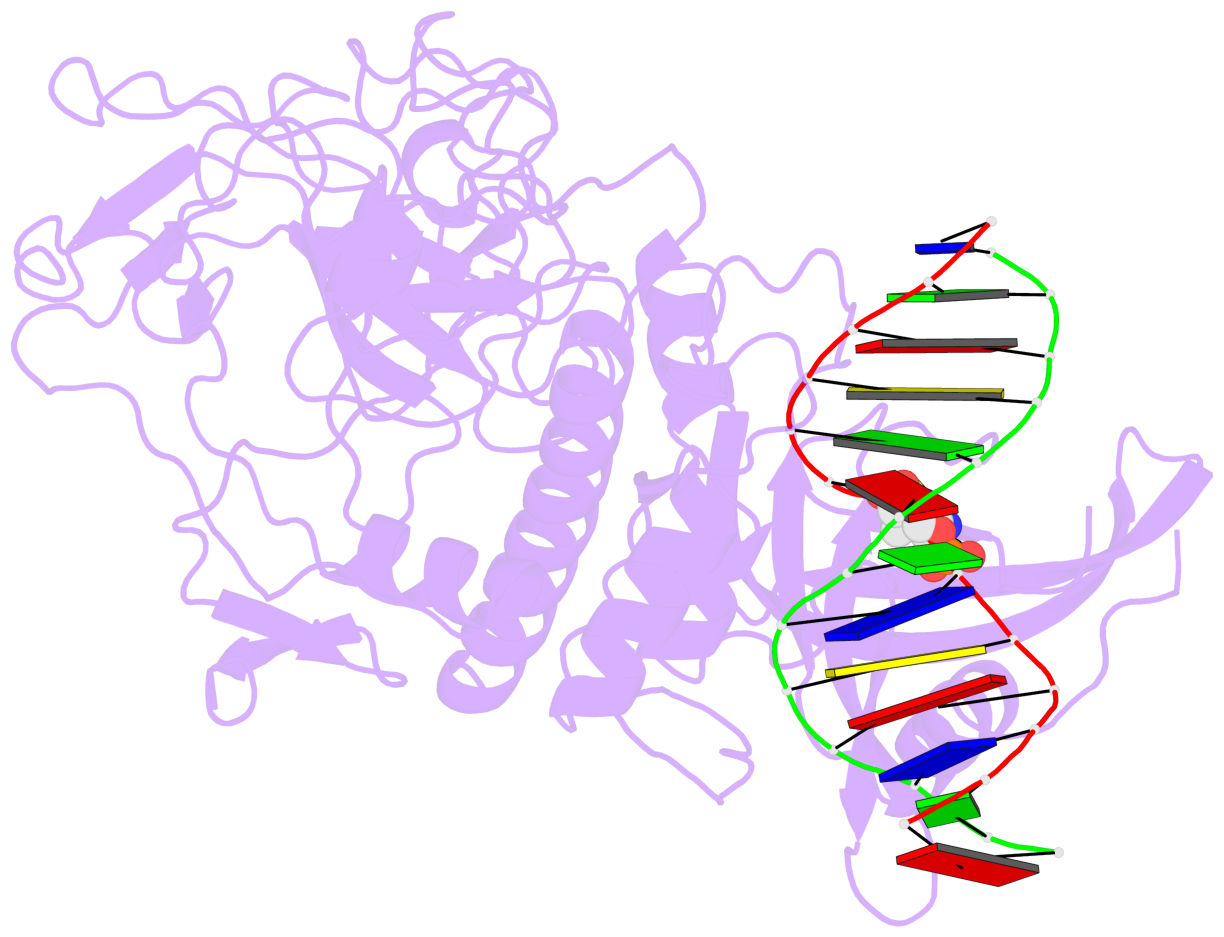
- The 5-methylcytosine group (PDB ligand '5CM') is shown in space-filling model,
with the methyl-carbon atom in black.
- Watson-Crick base pairs are represented as long rectangular blocks with the
minor-groove edge in black. Color code: A-T red, C-G yellow, G-C green, T-A blue.
- Protein is shown as cartoon in purple. DNA backbones are shown ribbon, colored code
by chain identifier.
- The block schematics were created with 3DNA-DSSR,
and images were rendered using PyMOL.
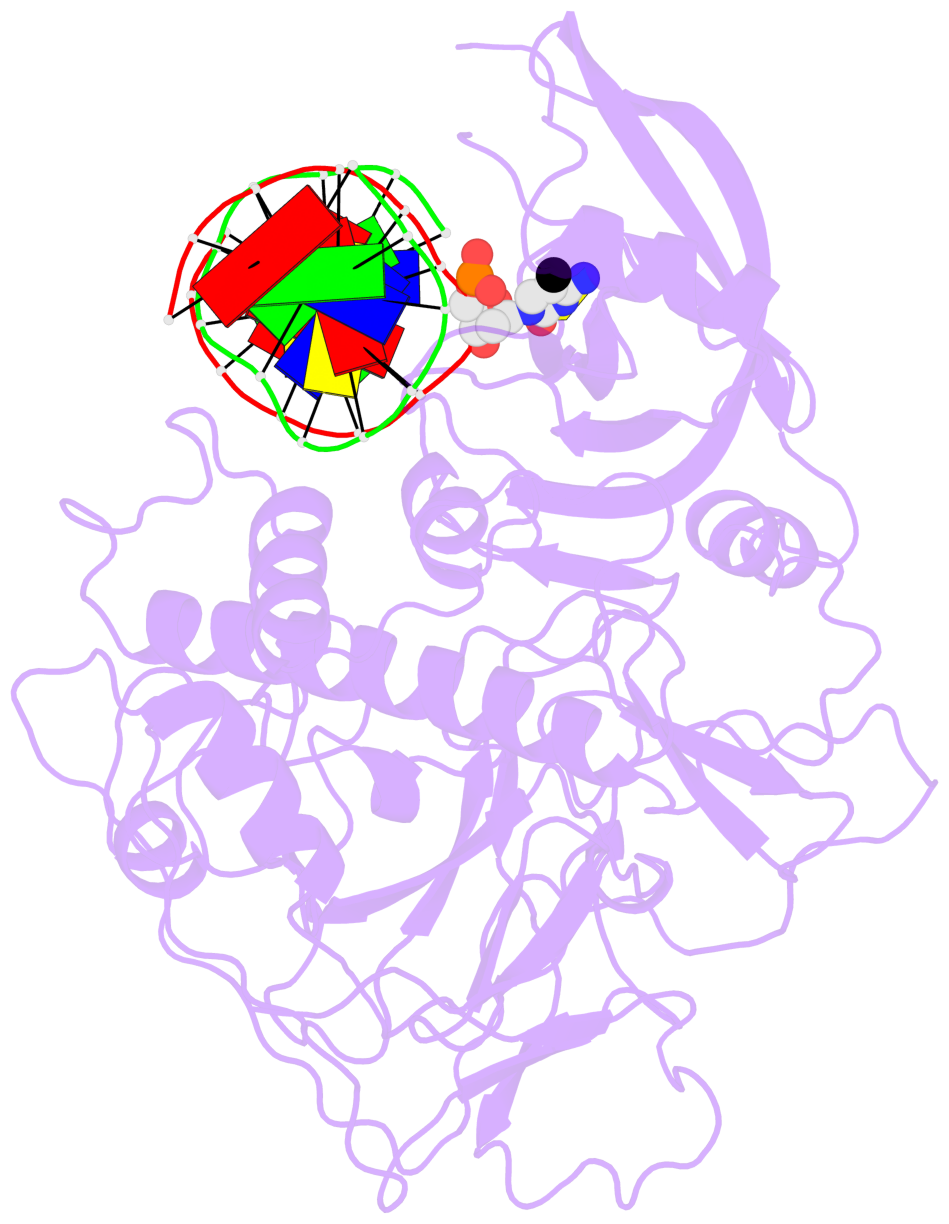
- Download the PyMOL session file corresponding to the top-left
image in the following panel.
- The contacts include paired nucleotides (mostly a G in G-C pairing), and
amino-acids within a 4.5-A distance cutoff to the base atoms of 5mC.
- The structure is oriented in the 'standard' base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
 |
No. 1 C.5CM8: download PDB file
for the 5mC entry
stacking-with-A.TYR380 stacking-with-A.TYR392 not-WC-paired not-in-duplex
|