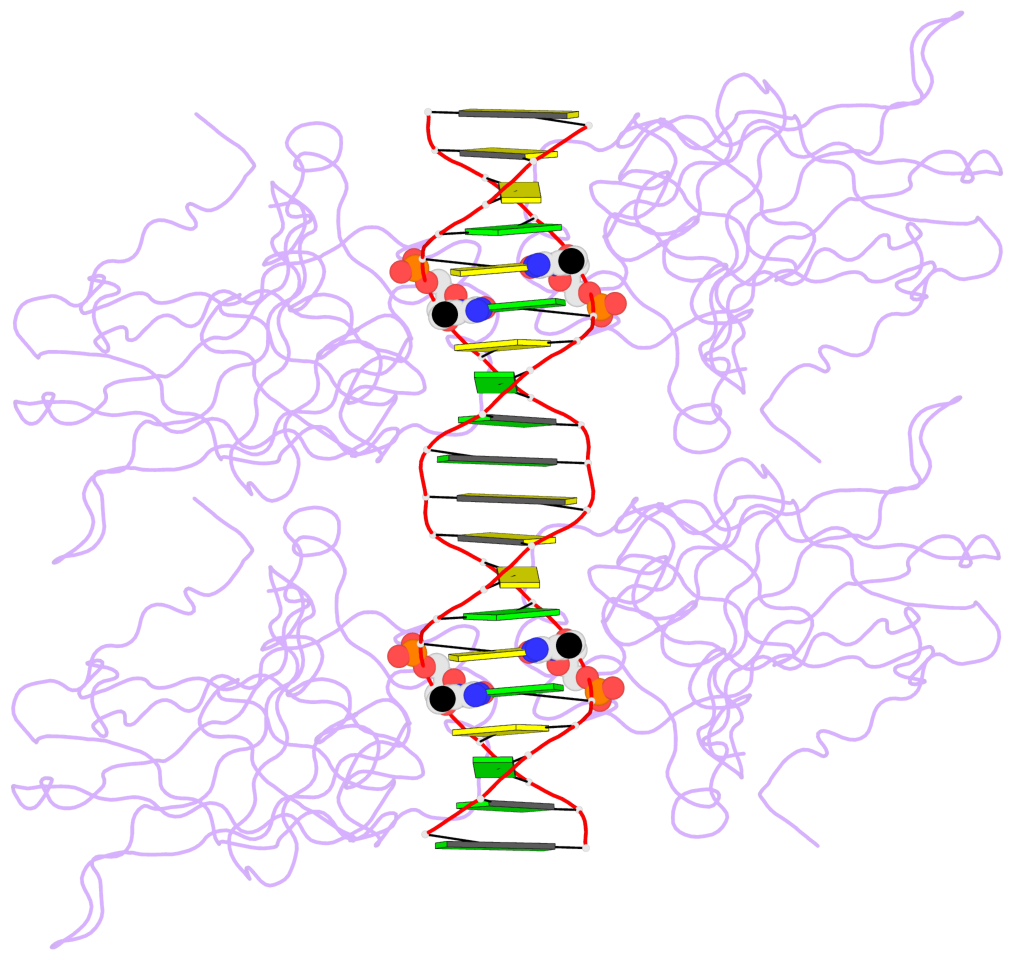
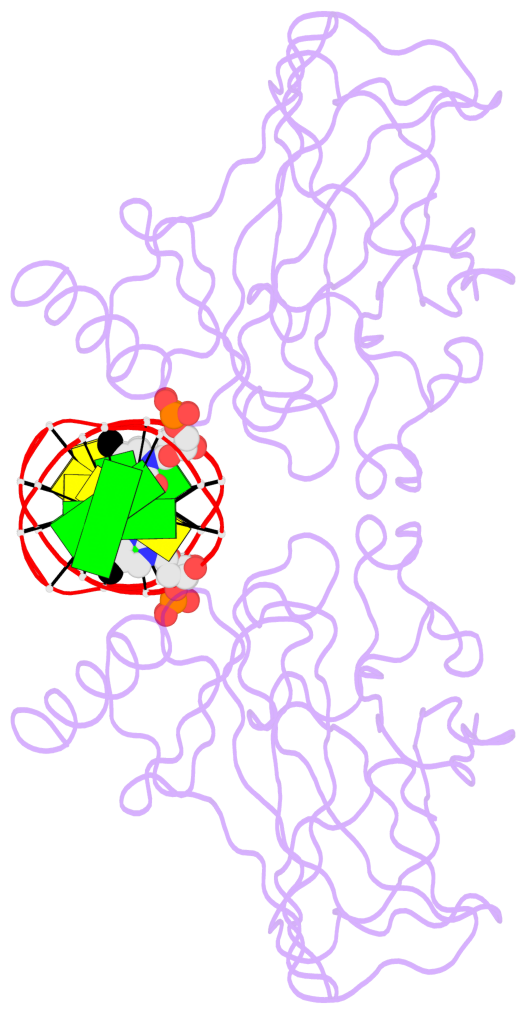
Last updated on 2019-09-30 by Xiang-Jun Lu <xiangjun@x3dna.org>.
The block schematics were created with DSSR and
rendered using PyMOL.
- PDB-id
- 5MCV
- Class
- transcription
- Method
- X-ray (1.6 Å)
- Summary
- New insights into the role of DNA shape on its recognition by p53 proteins (complex p53dbd-lwc1)
List of 4 5mC-amino acid contacts:
-
1:C.5CM6: stacking-with-1:A.ARG280 is-WC-paired is-in-duplex [+]:gcG/Cgc
-
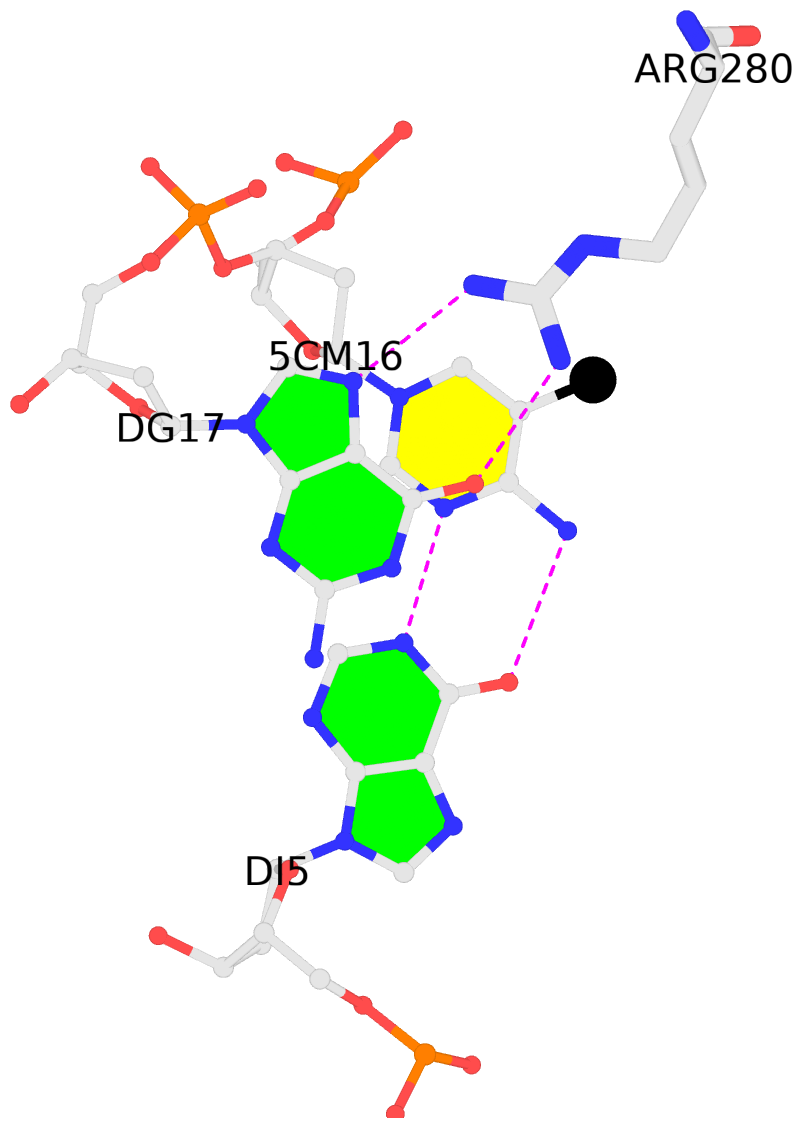
1:C.5CM16: stacking-with-1:B.ARG280 is-WC-paired is-in-duplex [+]:gcG/Cgc
-
2:C.5CM6: stacking-with-2:A.ARG280 is-WC-paired is-in-duplex [-]:Cgc/gcG
-
2:C.5CM16: stacking-with-2:B.ARG280 is-WC-paired is-in-duplex [-]:Cgc/gcG
direct SNAP output · DNAproDB 2.0
- Reference
- Golovenko, D., Brauning, B., Vyas, P., Haran, T.E., Rozenberg, H., Shakked, Z.: (2018) "New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins." Structure, 26, 1237-1250.e6.
- Abstract
- The tumor suppressor p53 acts as a transcription factor recognizing diverse DNA response elements (REs). Previous structural studies of p53-DNA complexes revealed non-canonical Hoogsteen geometry of A/T base pairs at conserved CATG motifs leading to changes in DNA shape and its interface with p53. To study the effects of DNA shape on binding characteristics, we designed REs with modified base pairs "locked" into either Hoogsteen or Watson-Crick form. Here we present crystal structures of these complexes and their thermodynamic and kinetic parameters, demonstrating that complexes with Hoogsteen base pairs are stabilized relative to those with all-Watson-Crick base pairs. CATG motifs are abundant in p53REs such as GADD45 and p53R2 related to cell-cycle arrest and DNA repair. The high-resolution structures of these complexes validate their propensity to adopt the unique Hoogsteen-induced structure, thus providing insights into the functional role of DNA shape and broadening the mechanisms that contribute to DNA recognition by proteins.
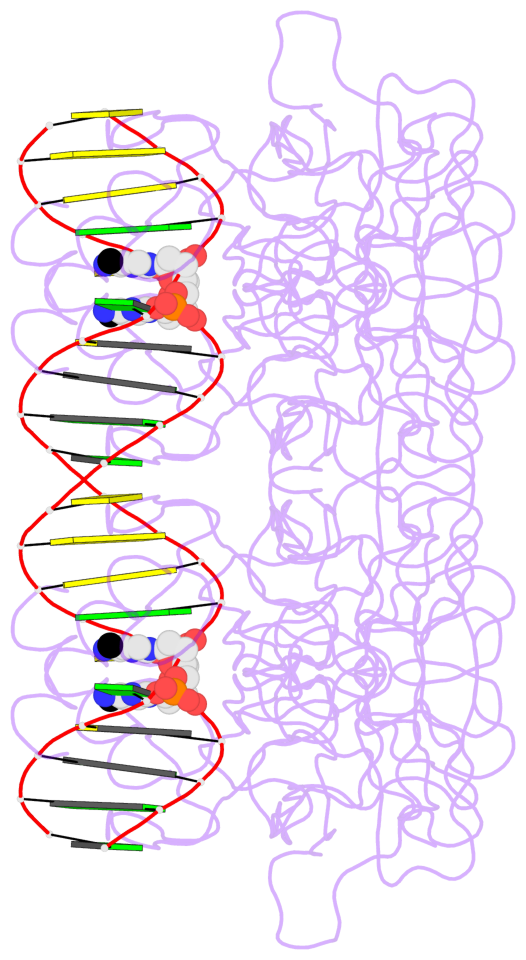
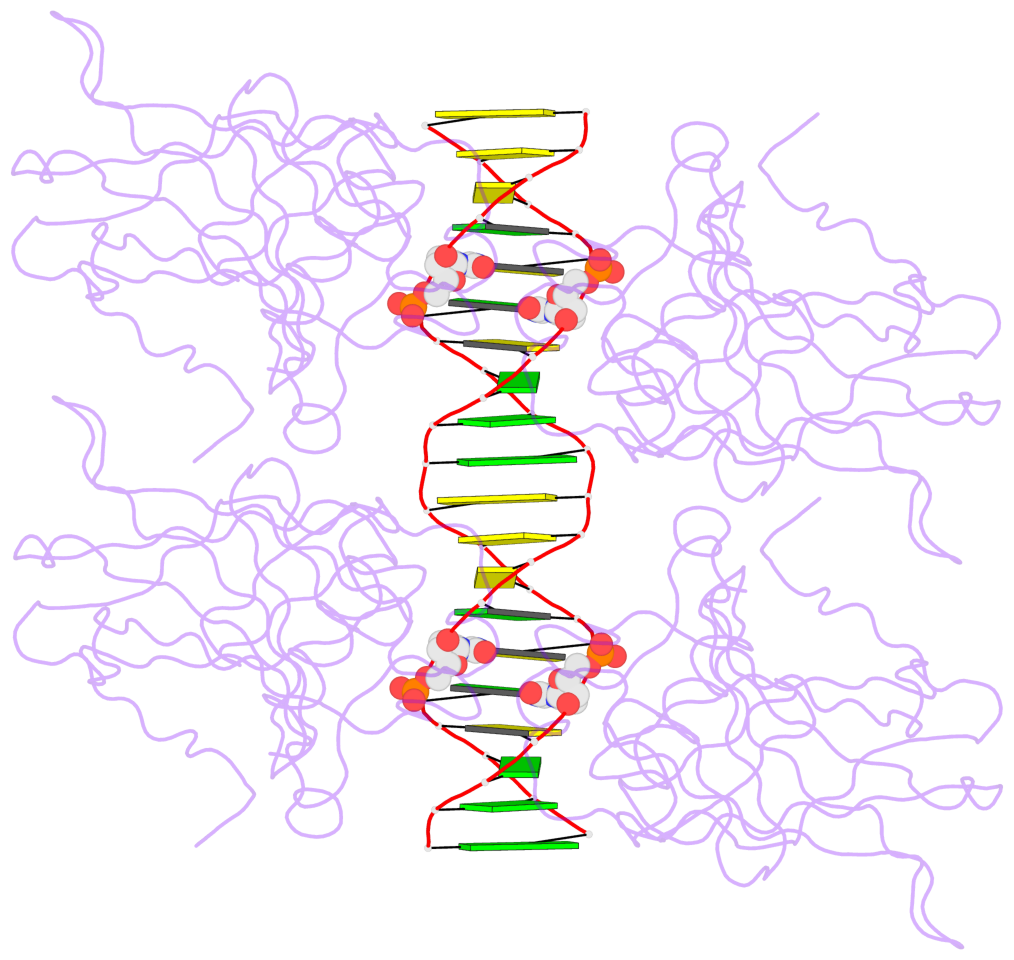
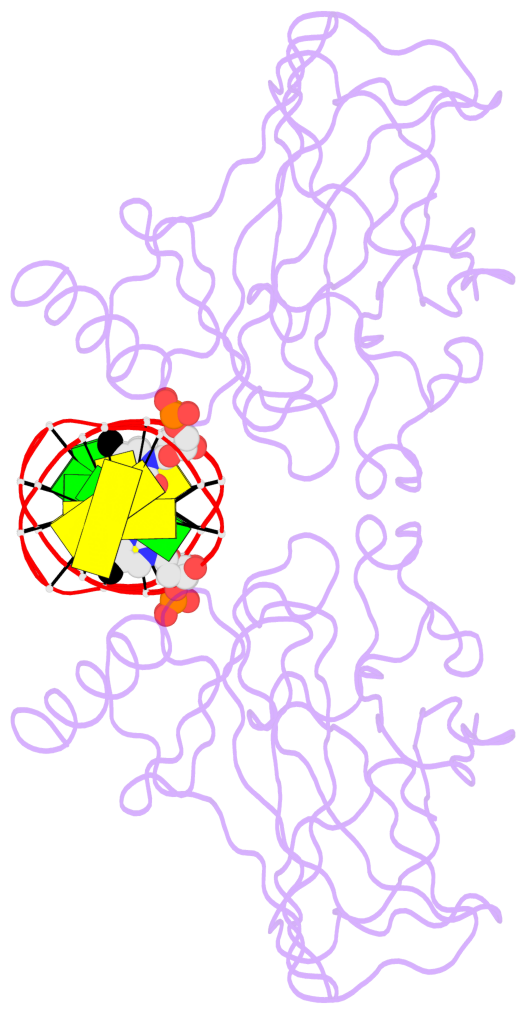
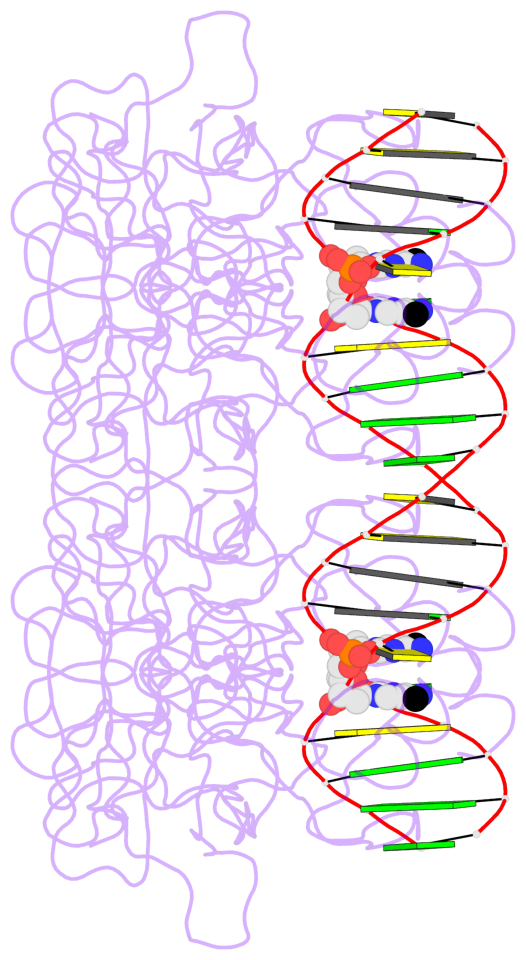
- The 5-methylcytosine group (PDB ligand '5CM') is shown in space-filling model,
with the methyl-carbon atom in black.
- Watson-Crick base pairs are represented as long rectangular blocks with the
minor-groove edge in black. Color code: A-T red, C-G yellow, G-C green, T-A blue.
- Protein is shown as cartoon in purple. DNA backbones are shown ribbon, colored code
by chain identifier.
- The block schematics were created with 3DNA-DSSR,
and images were rendered using PyMOL.
- Download the PyMOL session file corresponding to the top-left
image in the following panel.
- The contacts include paired nucleotides (mostly a G in G-C pairing), and
amino-acids within a 4.5-A distance cutoff to the base atoms of 5mC.
- The structure is oriented in the 'standard' base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
 |
No. 1 1:C.5CM6: download PDB file
for the 5mC entry
stacking-with-1:A.ARG280 is-WC-paired is-in-duplex [+]:gcG/Cgc
|
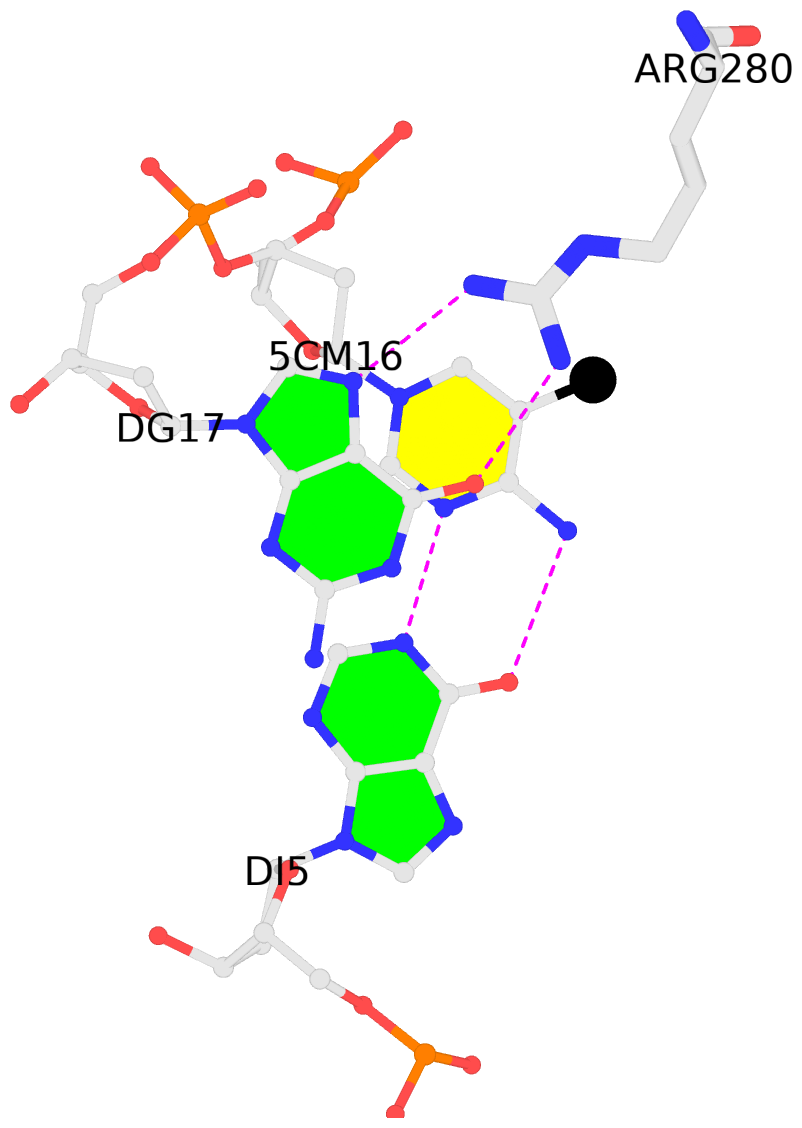 |
No. 2 1:C.5CM16: download PDB file
for the 5mC entry
stacking-with-1:B.ARG280 is-WC-paired is-in-duplex [+]:gcG/Cgc
|
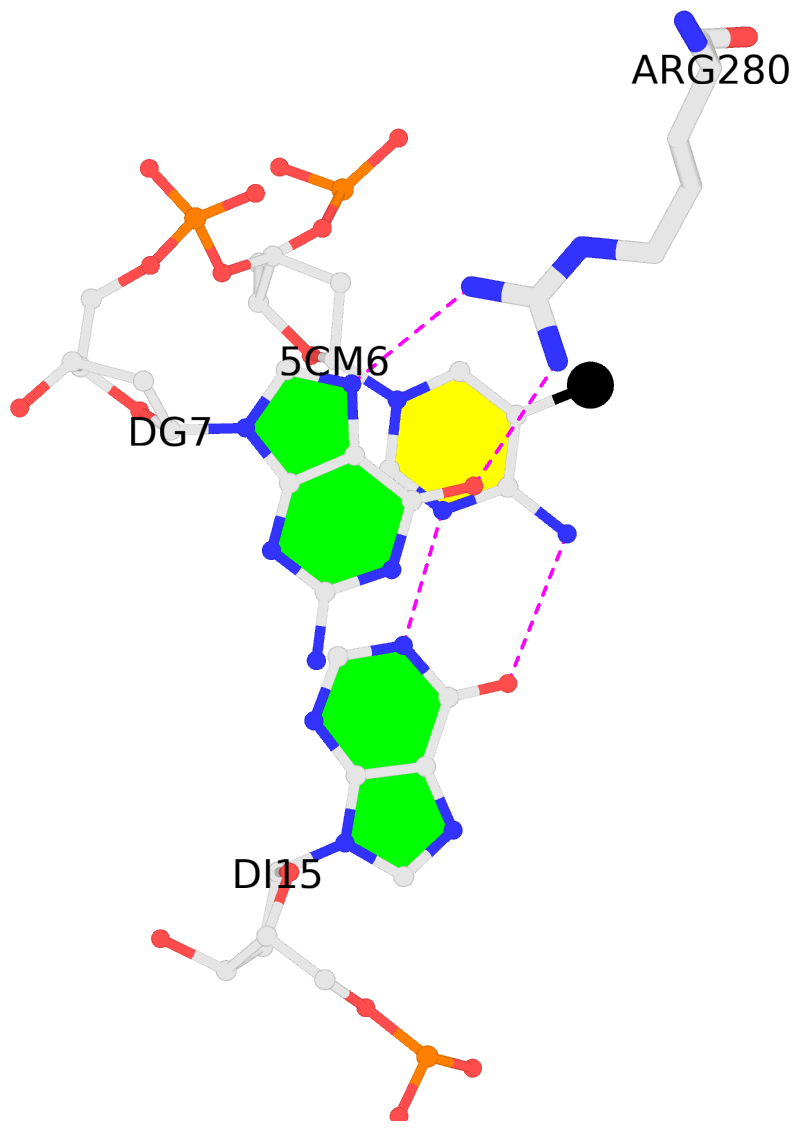 |
No. 3 2:C.5CM6: download PDB file
for the 5mC entry
stacking-with-2:A.ARG280 is-WC-paired is-in-duplex [-]:Cgc/gcG
|
 |
No. 4 2:C.5CM16: download PDB file
for the 5mC entry
stacking-with-2:B.ARG280 is-WC-paired is-in-duplex [-]:Cgc/gcG
|