Last updated on 2019-09-30 by Xiang-Jun Lu <xiangjun@x3dna.org>.
The block schematics were created with DSSR and
rendered using PyMOL.
- PDB-id
- 4X9J
- Class
- transcription regulator-DNA
- Method
- X-ray (1.41 Å)
- Summary
- Egr-1 with doubly methylated DNA
List of 4 5mC-amino acid contacts:
-
B.5CM3: stacking-with-A.ARG174 is-WC-paired is-in-duplex [+]:GcG/cGC
-
B.5CM9: stacking-with-A.ARG118 is-WC-paired is-in-duplex [+]:GcG/cGC
-
C.5CM53: other-contacts is-WC-paired is-in-duplex [-]:cGT/AcG
-
C.5CM59: other-contacts is-WC-paired is-in-duplex [-]:cGT/AcG
direct SNAP output · DNAproDB 2.0
- Reference
- Zandarashvili, L., White, M.A., Esadze, A., Iwahara, J.: (2015) "Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1." Febs Lett., 589, 1748-1753.
- Abstract
- The inducible transcription factor Egr-1 binds specifically to 9-bp target sequences containing two CpG sites that can potentially be methylated at four cytosine bases. Although it appears that complete CpG methylation would make an unfavorable steric clash in the previous crystal structures of the complexes with unmethylated or partially methylated DNA, our affinity data suggest that DNA recognition by Egr-1 is insensitive to CpG methylation. We have determined, at a 1.4-Å resolution, the crystal structure of the Egr-1 zinc-finger complex with completely methylated target DNA. Structural comparison of the three different methylation states reveals why Egr-1 can recognize the target sequences regardless of CpG methylation.
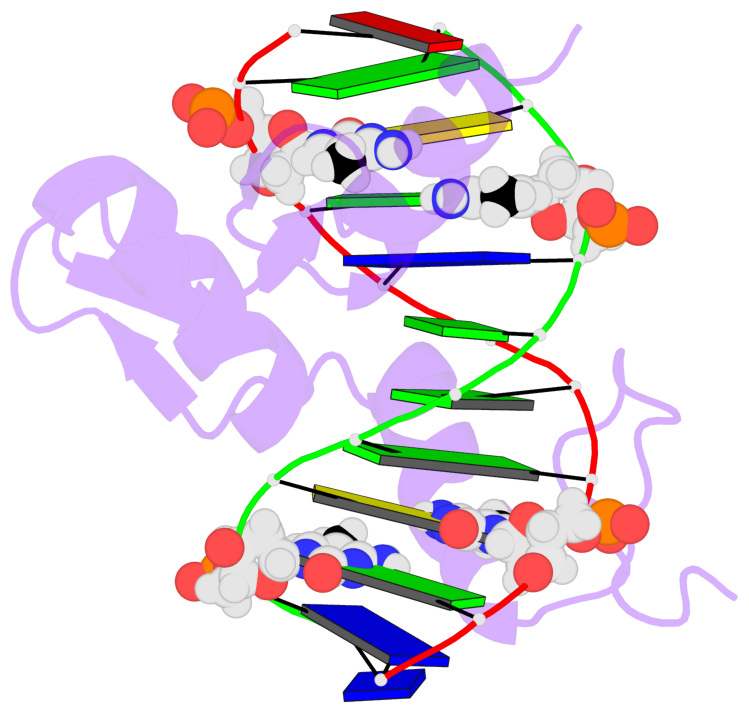
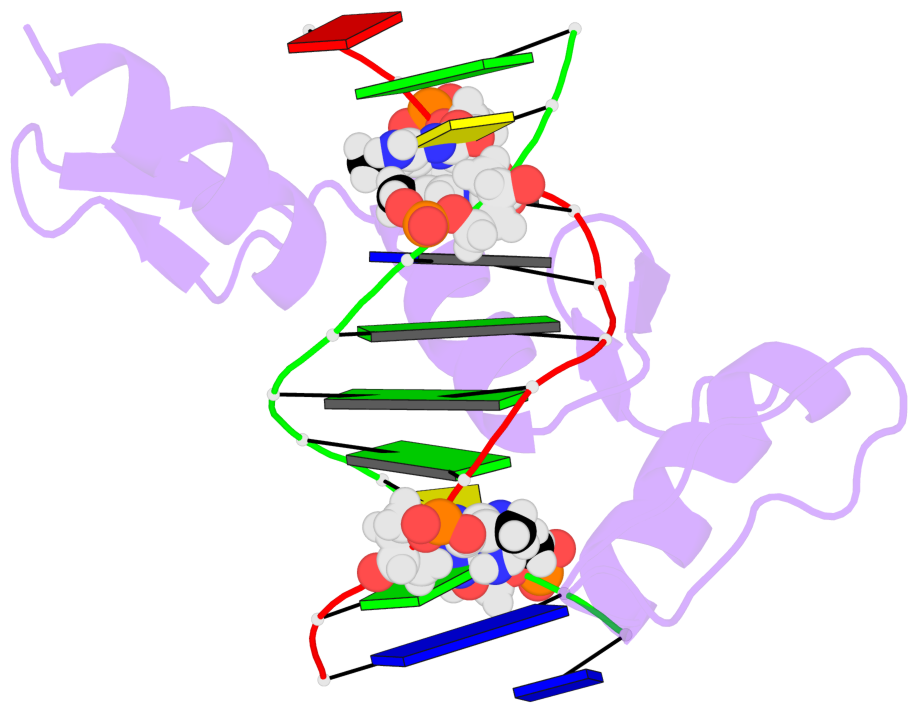
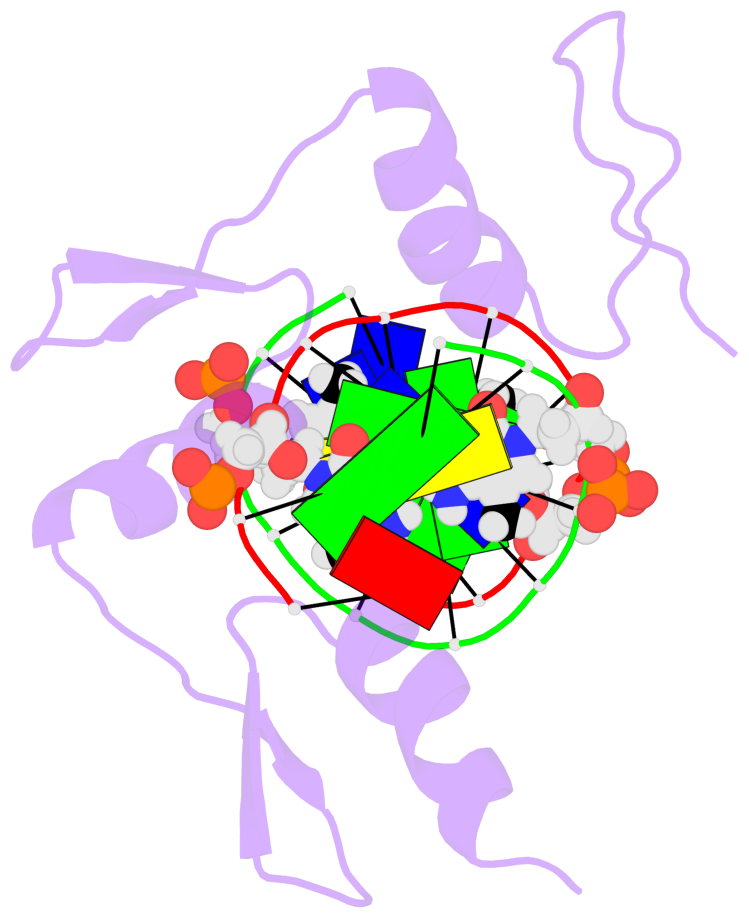
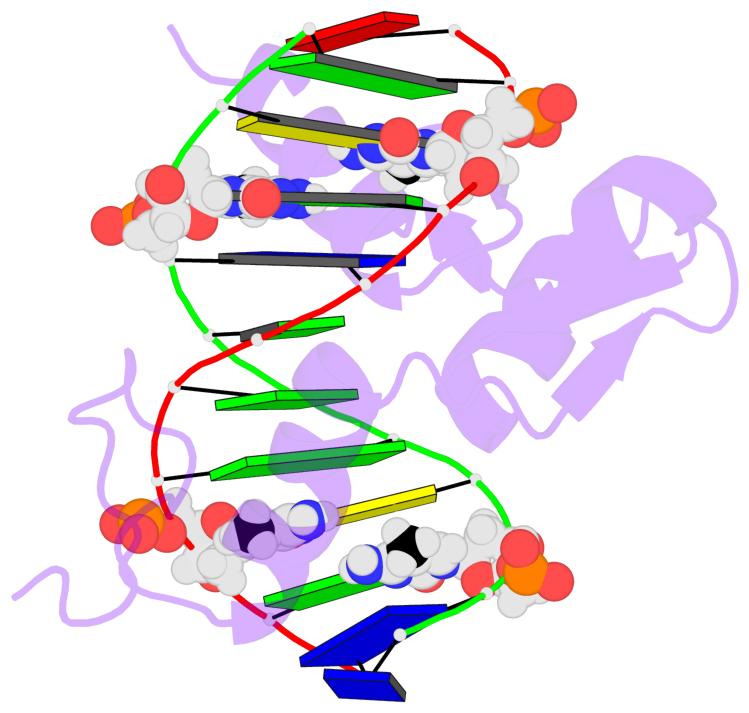
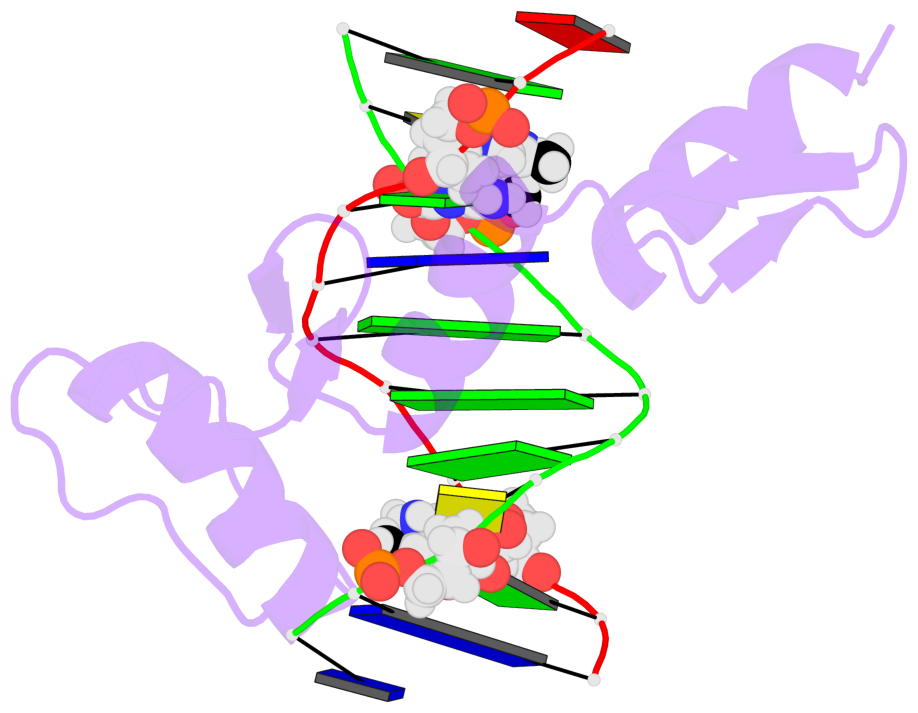
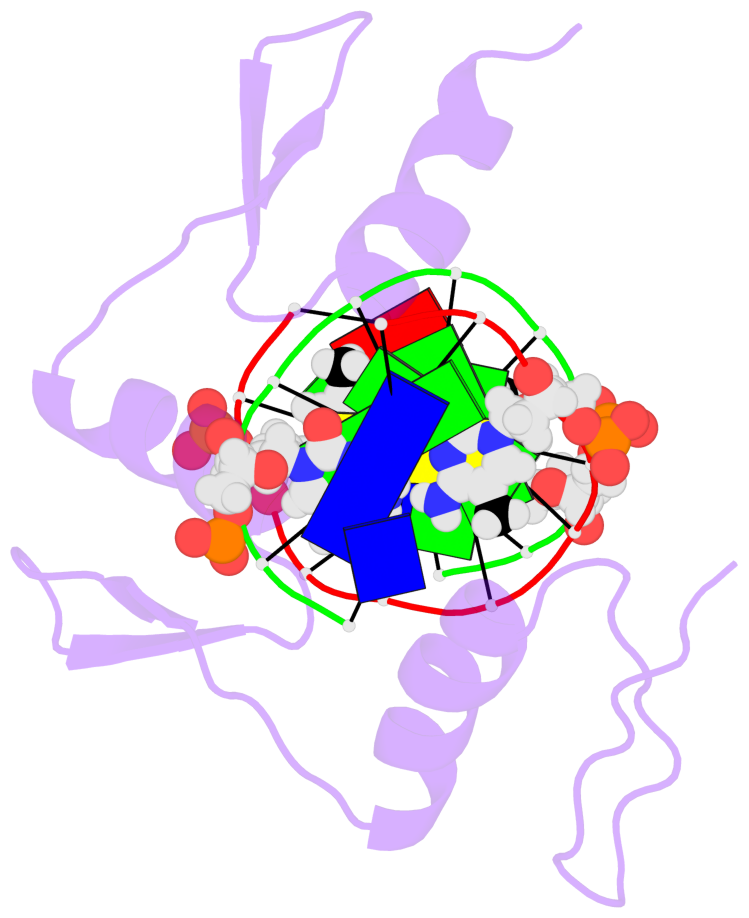
- The 5-methylcytosine group (PDB ligand '5CM') is shown in space-filling model,
with the methyl-carbon atom in black.
- Watson-Crick base pairs are represented as long rectangular blocks with the
minor-groove edge in black. Color code: A-T red, C-G yellow, G-C green, T-A blue.
- Protein is shown as cartoon in purple. DNA backbones are shown ribbon, colored code
by chain identifier.
- The block schematics were created with 3DNA-DSSR,
and images were rendered using PyMOL.
- Download the PyMOL session file corresponding to the top-left
image in the following panel.
- The contacts include paired nucleotides (mostly a G in G-C pairing), and
amino-acids within a 4.5-A distance cutoff to the base atoms of 5mC.
- The structure is oriented in the 'standard' base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
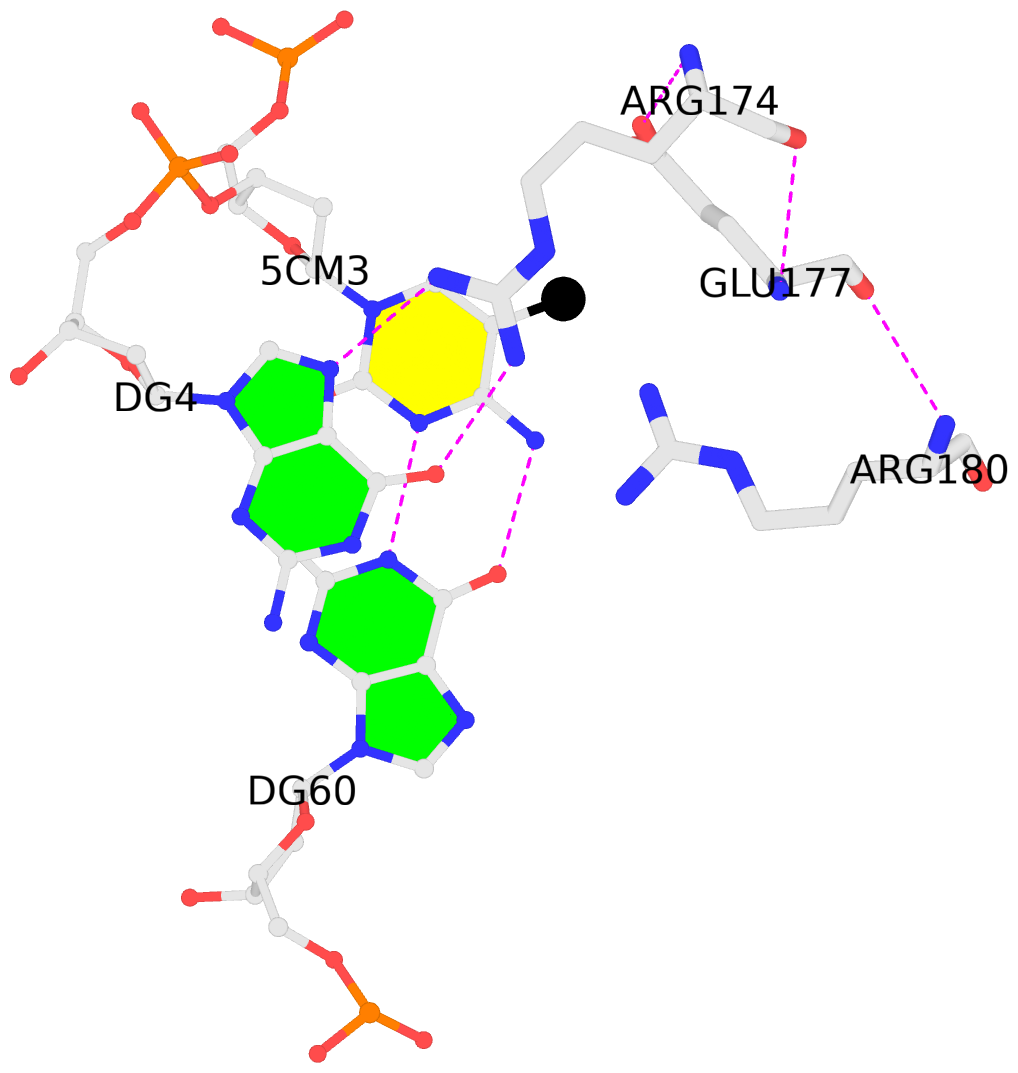 |
No. 1 B.5CM3: download PDB file
for the 5mC entry
stacking-with-A.ARG174 is-WC-paired is-in-duplex [+]:GcG/cGC
|
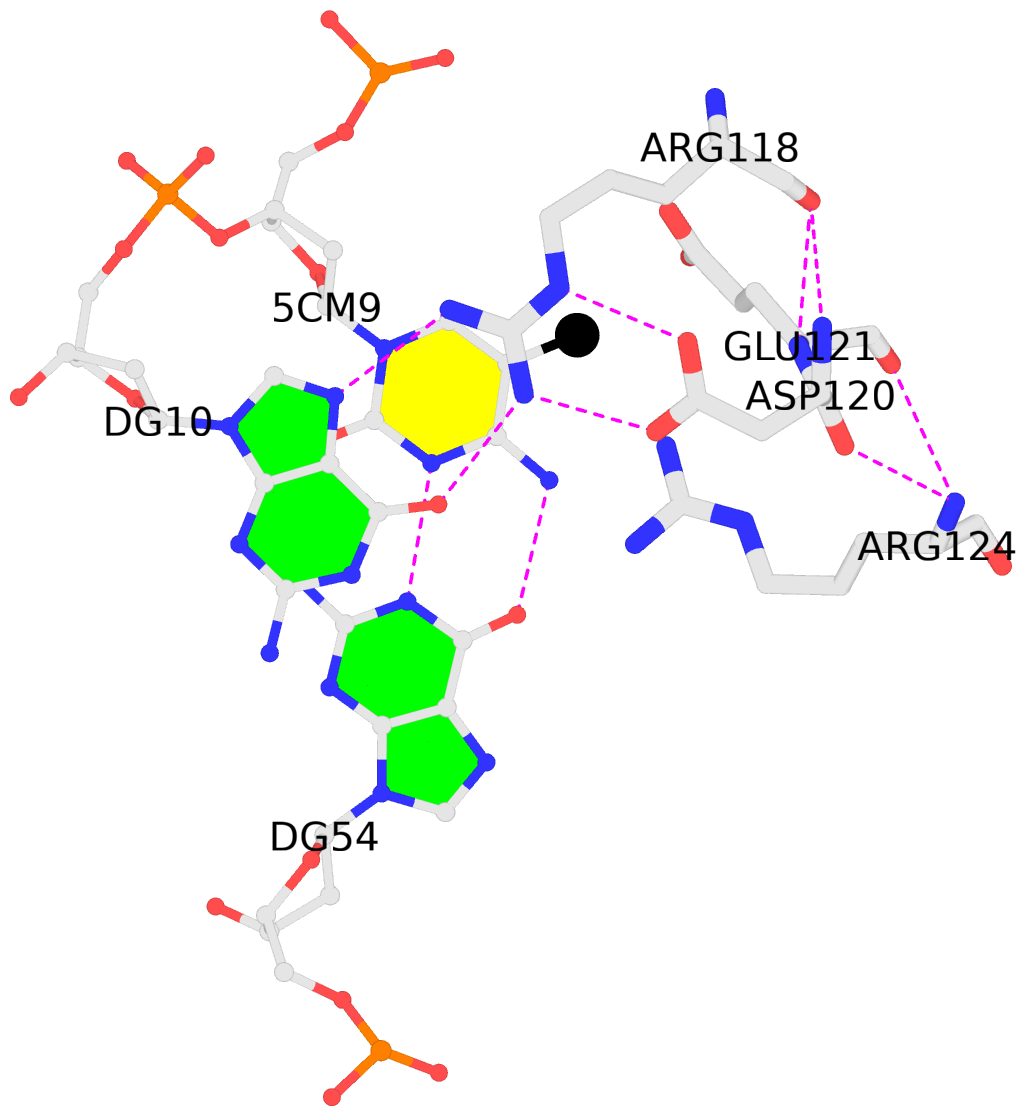 |
No. 2 B.5CM9: download PDB file
for the 5mC entry
stacking-with-A.ARG118 is-WC-paired is-in-duplex [+]:GcG/cGC
|
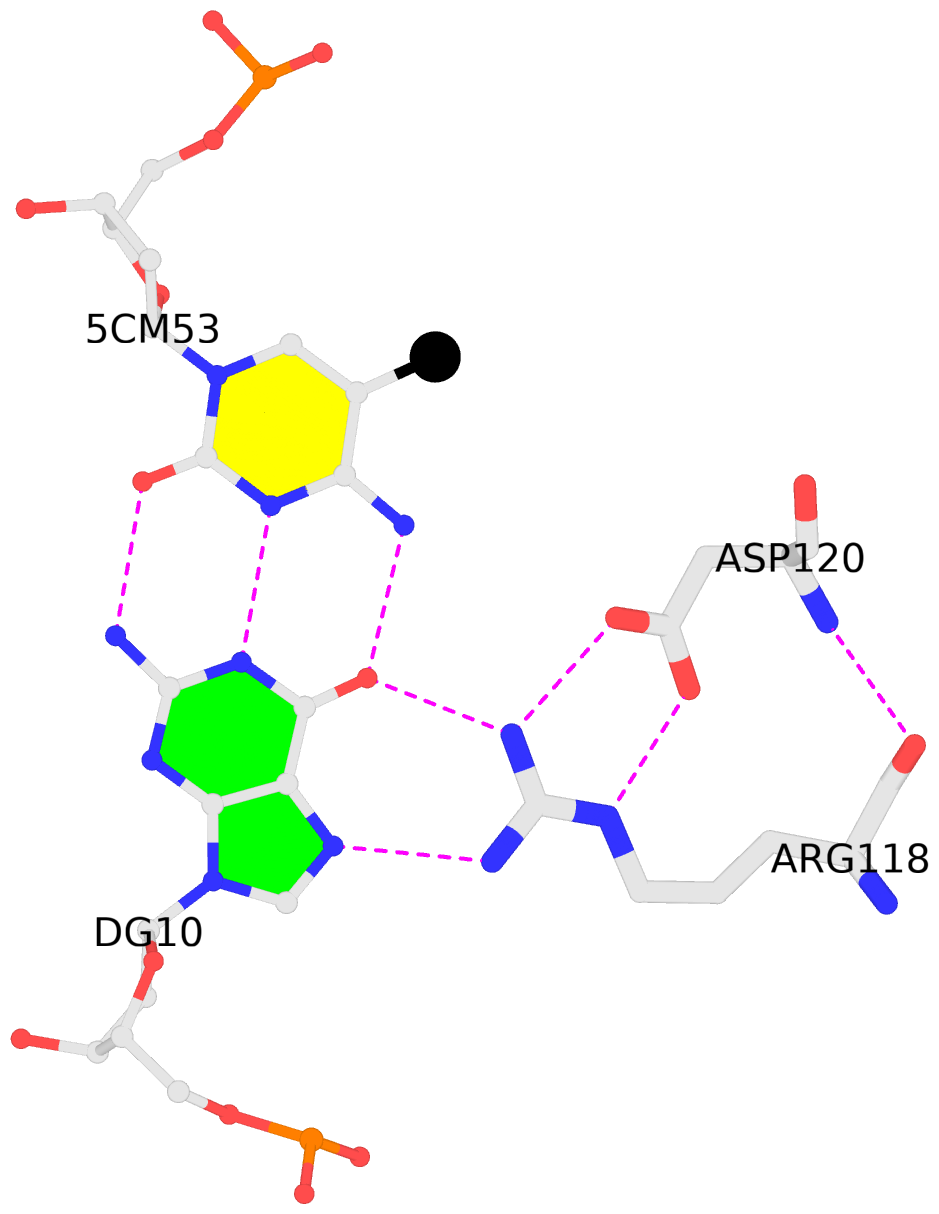 |
No. 3 C.5CM53: download PDB file
for the 5mC entry
other-contacts is-WC-paired is-in-duplex [-]:cGT/AcG
|
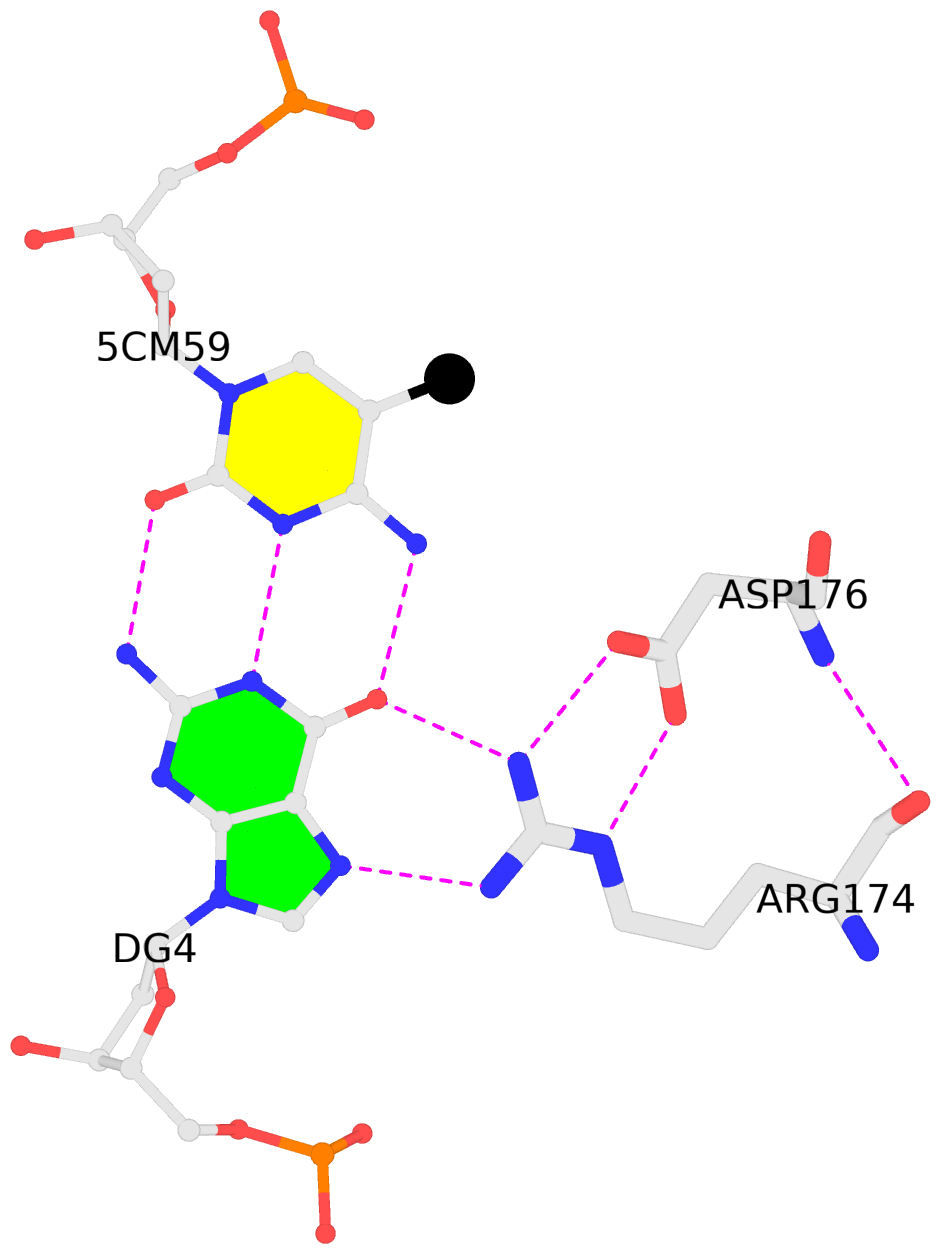 |
No. 4 C.5CM59: download PDB file
for the 5mC entry
other-contacts is-WC-paired is-in-duplex [-]:cGT/AcG
|