5mC interactions in PDB entry 4HP1 auto-curated with SNAP
Last updated on 2019-09-30 by Xiang-Jun Lu <xiangjun@x3dna.org>. The block schematics were created with DSSR and rendered using PyMOL.
Summary information and primary citation [schematics · contacts · top · homepage · tutorial]
- PDB-id
- 4HP1
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.25 Å)
- Summary
- Crystal structure of tet3 in complex with a non-cpg dsDNA
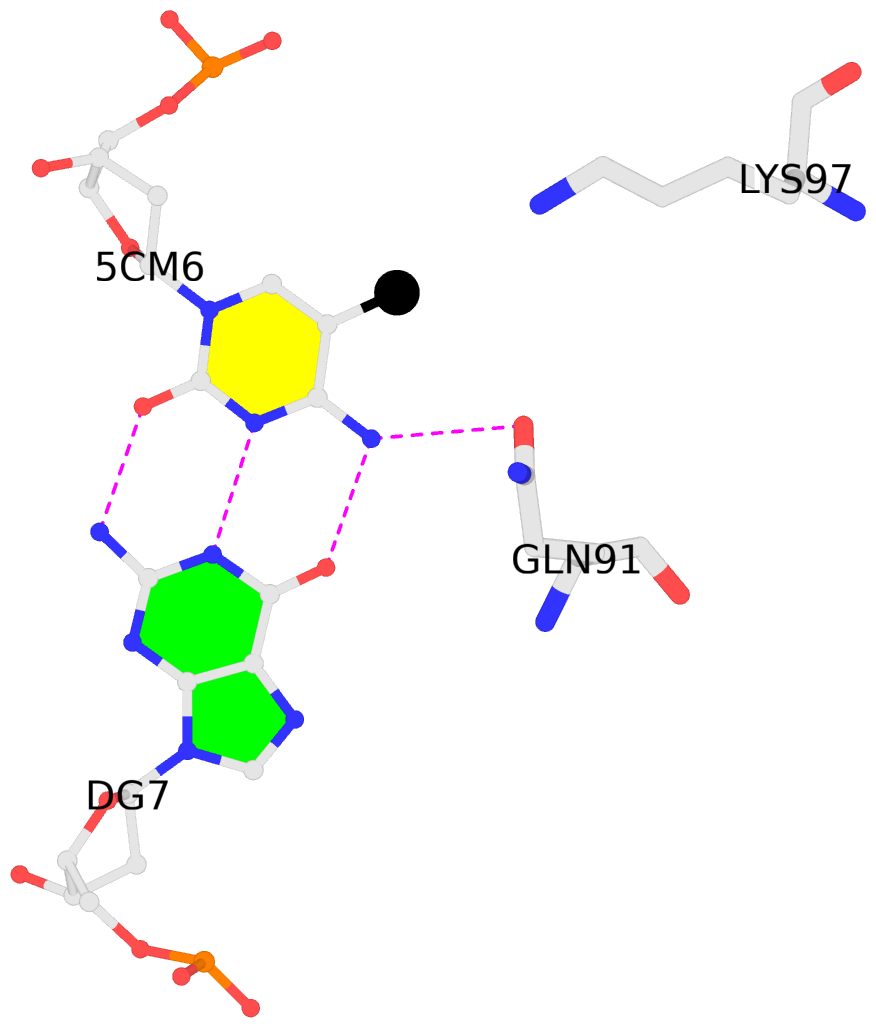
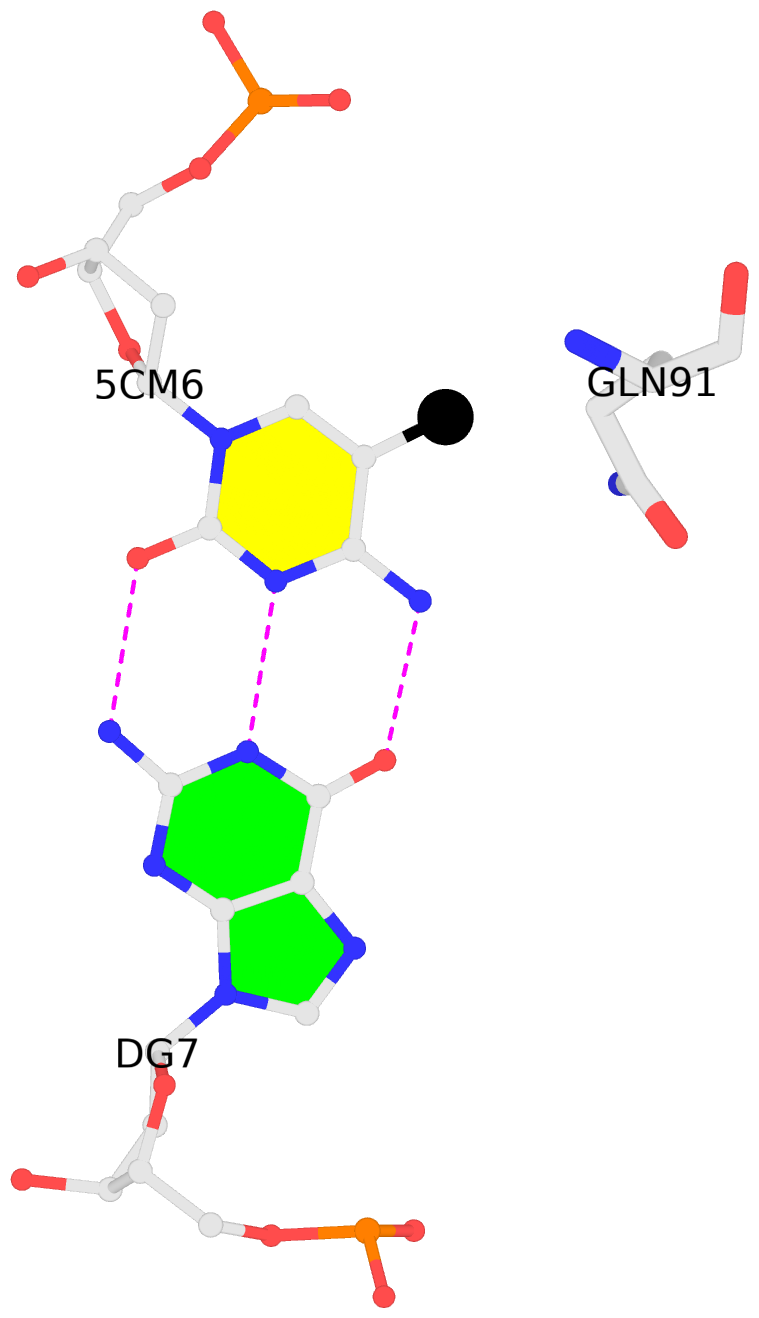
List of 2 5mC-amino acid contacts:-
A.5CM6: other-contacts is-WC-paired is-in-duplex [+]:CcG/cGG
-
B.5CM6: other-contacts is-WC-paired is-in-duplex [-]:cGG/CcG
-
- Reference
- Xu, Y., Xu, C., Kato, A., Tempel, W., Abreu, J.G., Bian, C., Hu, Y., Hu, D., Zhao, B., Cerovina, T., Diao, J., Wu, F., He, H.H., Cui, Q., Clark, E., Ma, C., Barbara, A., Veenstra, G.J., Xu, G., Kaiser, U.B., Liu, X.S., Sugrue, S.P., He, X., Min, J., Kato, Y., Shi, Y.G.: (2012) "Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development." Cell, 151, 1200-1213.
- Abstract
- Ten-Eleven Translocation (Tet) family of dioxygenases dynamically regulates DNA methylation and has been implicated in cell lineage differentiation and oncogenesis. Yet their functions and mechanisms of action in gene regulation and embryonic development are largely unknown. Here, we report that Xenopus Tet3 plays an essential role in early eye and neural development by directly regulating a set of key developmental genes. Tet3 is an active 5mC hydroxylase regulating the 5mC/5hmC status at target gene promoters. Biochemical and structural studies further demonstrate that the Tet3 CXXC domain is critical for specific Tet3 targeting. Finally, we show that the enzymatic activity and CXXC domain are both crucial for Tet3's biological function. Together, these findings define Tet3 as a transcription regulator and reveal a molecular mechanism by which the 5mC hydroxylase and DNA binding activities of Tet3 cooperate to control target gene expression and embryonic development.
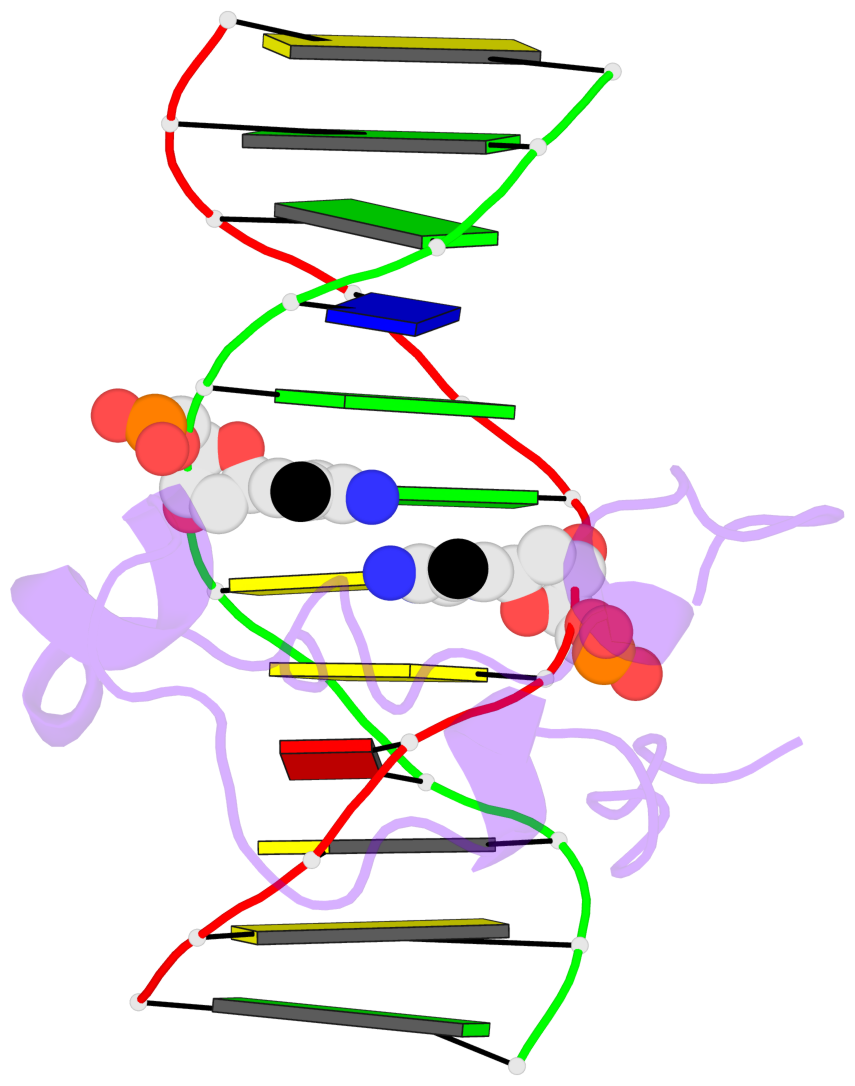
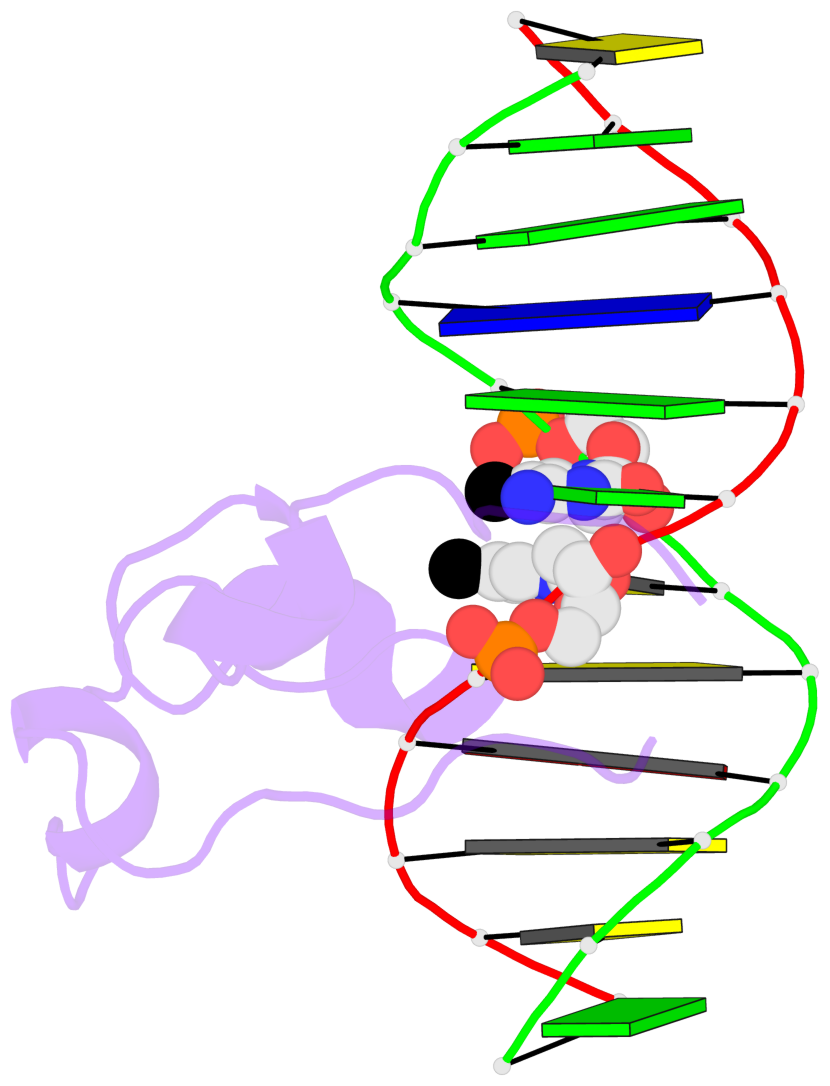
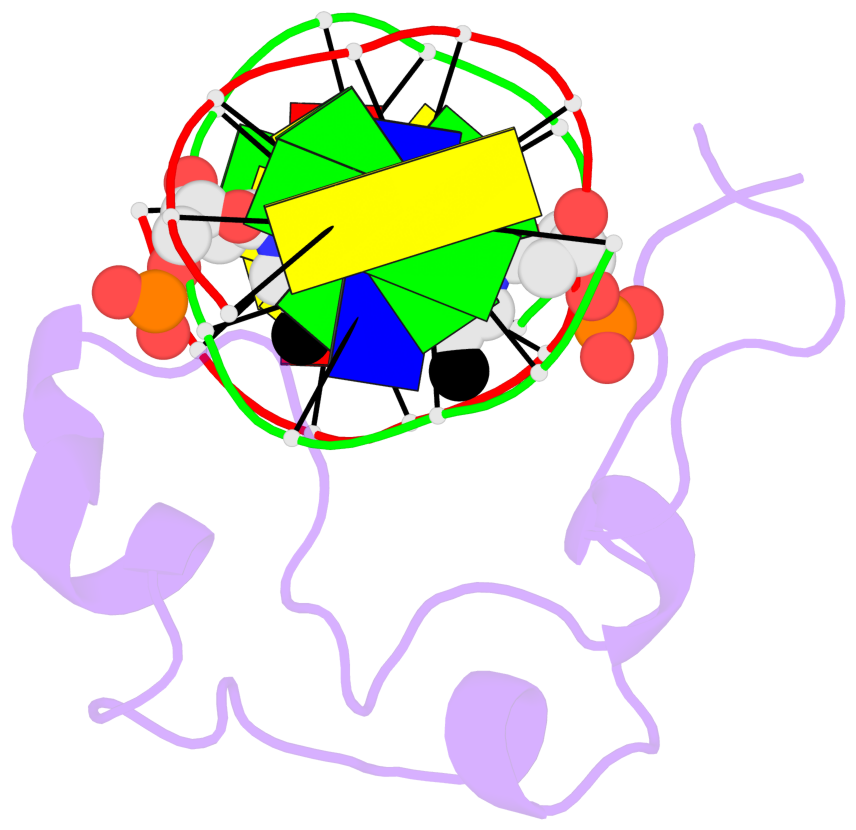
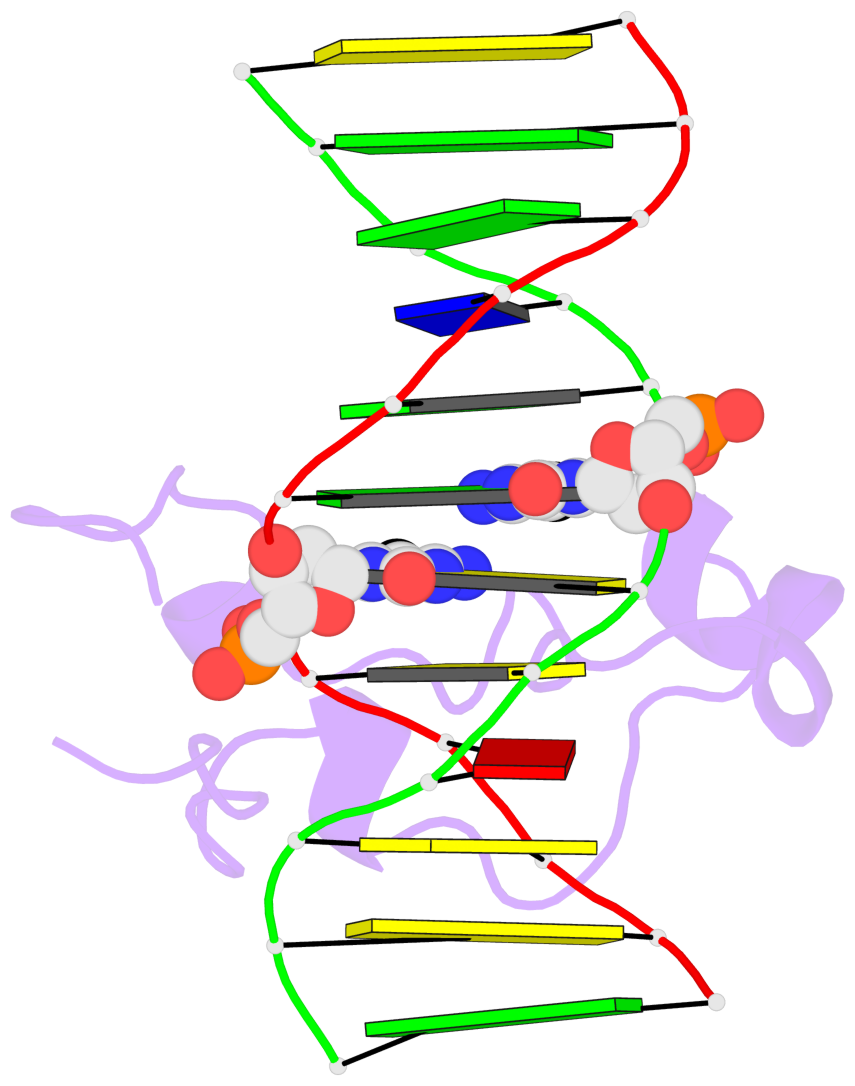
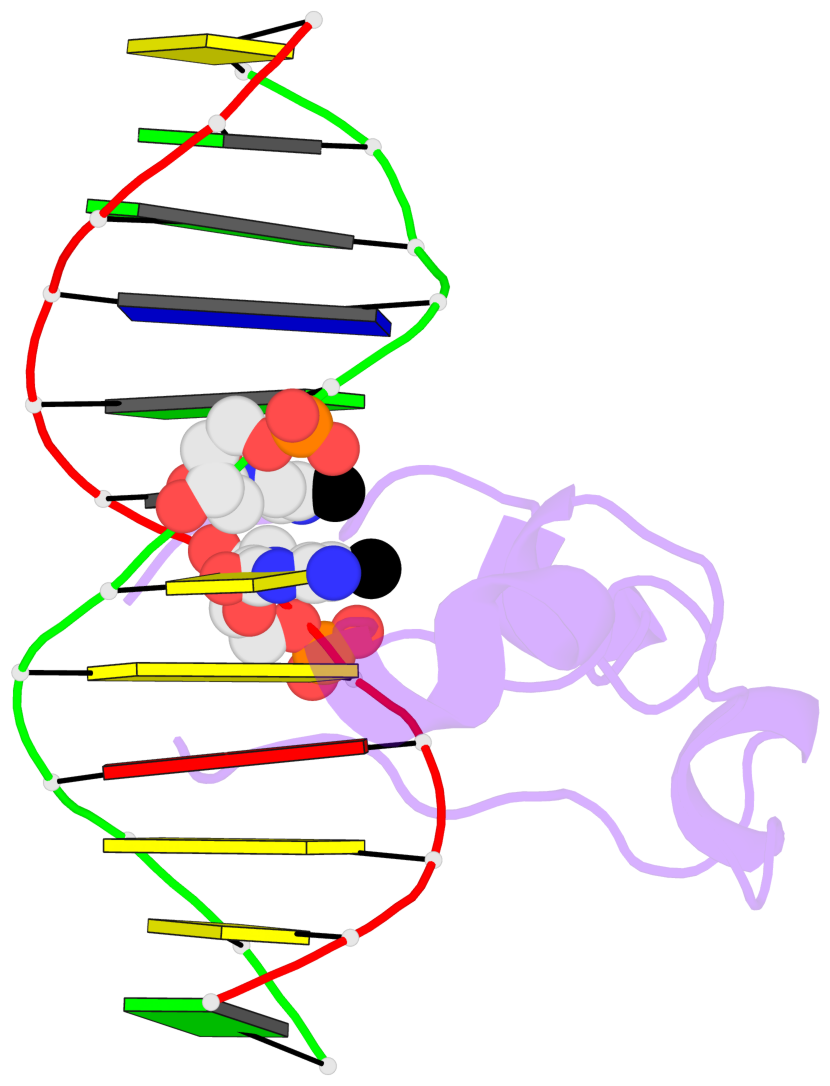
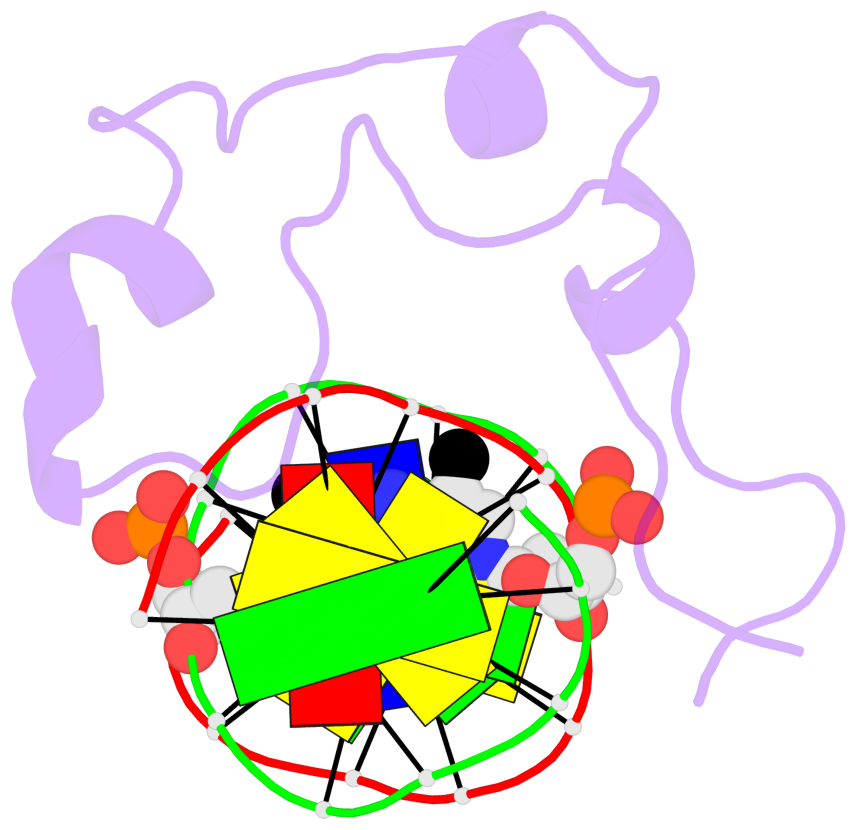
Base-block schematics in six views [summary · contacts · top · homepage · tutorial]
- The 5-methylcytosine group (PDB ligand '5CM') is shown in space-filling model, with the methyl-carbon atom in black.
- Watson-Crick base pairs are represented as long rectangular blocks with the minor-groove edge in black. Color code: A-T red, C-G yellow, G-C green, T-A blue.
- Protein is shown as cartoon in purple. DNA backbones are shown ribbon, colored code by chain identifier.
- The block schematics were created with 3DNA-DSSR, and images were rendered using PyMOL.
- Download the PyMOL session file corresponding to the top-left image in the following panel.
 |
 |
 |
 |
 |
 |
List of 2 5mC-amino acid contacts [summary · schematics · top · homepage · tutorial]
- The contacts include paired nucleotides (mostly a G in G-C pairing), and amino-acids within a 4.5-A distance cutoff to the base atoms of 5mC.
- The structure is oriented in the 'standard' base reference frame of 5mC, allowing for easy comparison and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.