5mC interactions in PDB entry 4GZN auto-curated with SNAP
Last updated on 2019-09-30 by Xiang-Jun Lu <xiangjun@x3dna.org>. The block schematics were created with DSSR and rendered using PyMOL.
Summary information and primary citation [schematics · contacts · top · homepage · tutorial]
- PDB-id
- 4GZN
- Class
- transcription-DNA
- Method
- X-ray (0.99 Å)
- Summary
- Mouse zfp57 zinc fingers in complex with methylated DNA
List of 2 5mC-amino acid contacts:-
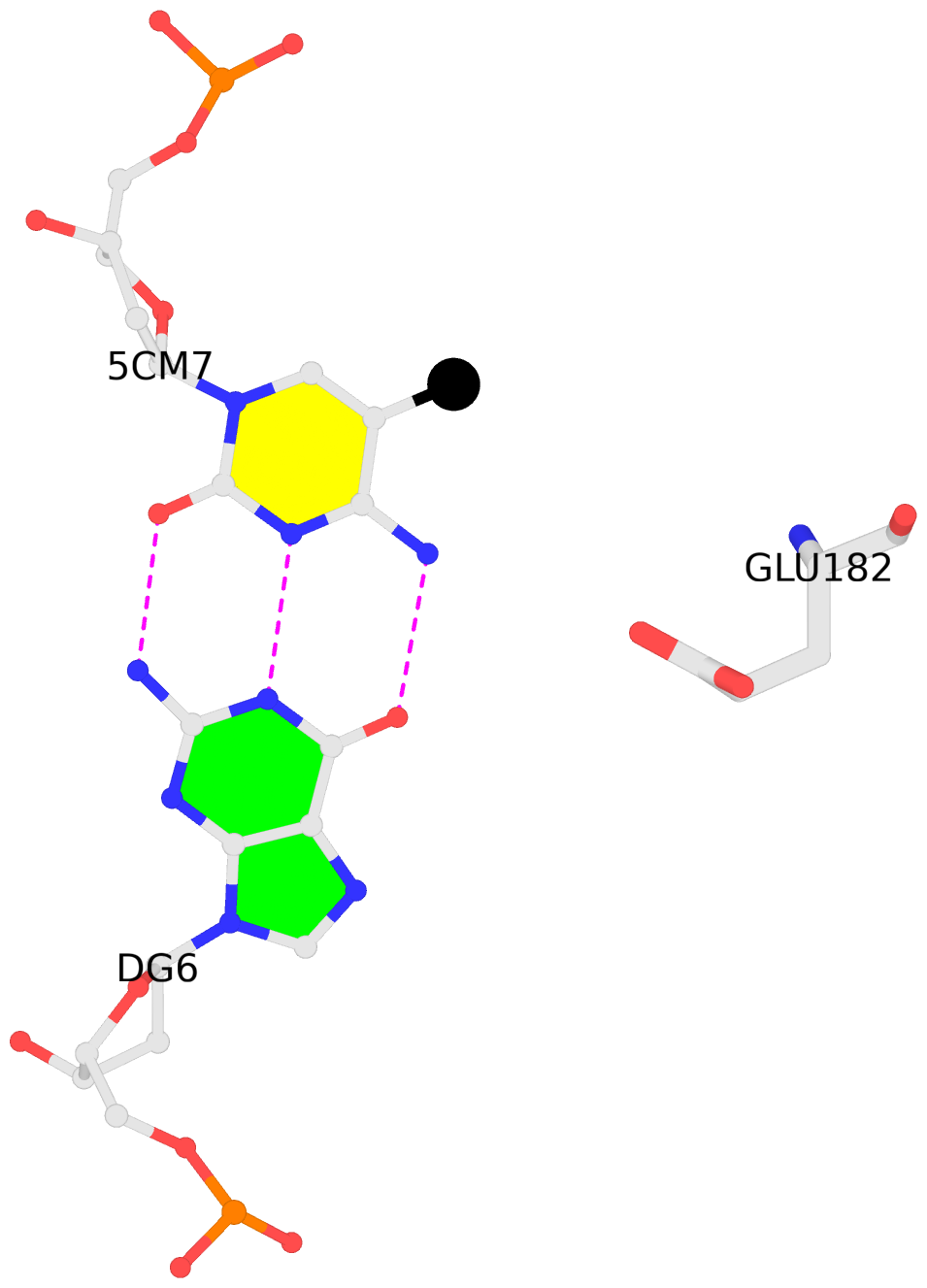
A.5CM7: other-contacts is-WC-paired is-in-duplex [+]:CcG/cGG
-
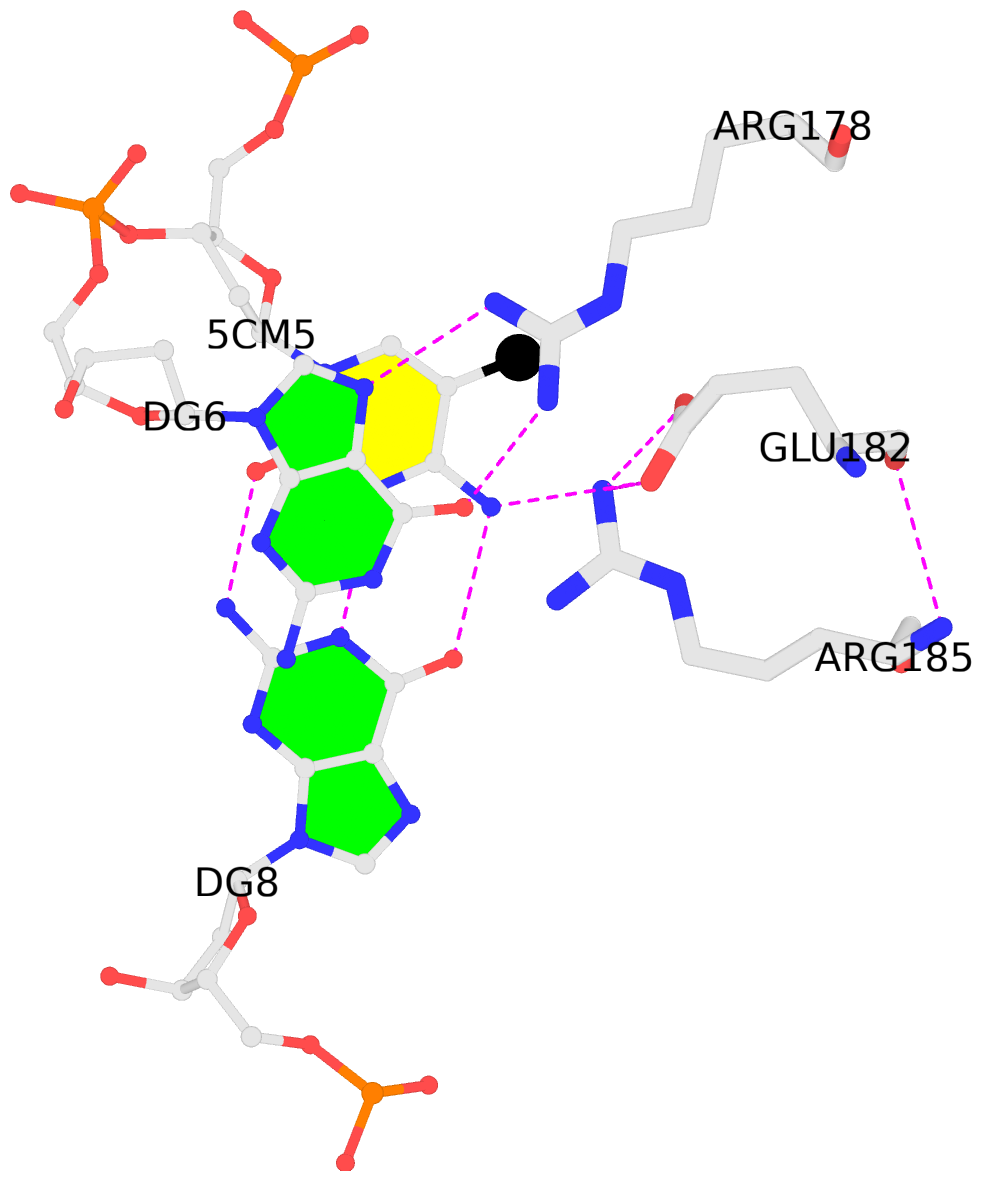
B.5CM5: stacking-with-C.ARG178 is-WC-paired is-in-duplex [-]:cGC/GcG
-
- Reference
- Liu, Y., Toh, H., Sasaki, H., Zhang, X., Cheng, X.: (2012) "An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence." Genes Dev., 26, 2374-2379.
- Abstract
- Zinc finger transcription factor Zfp57 recognizes the methylated CpG within the TGCCGC element. We determined the structure of the DNA-binding domain of Zfp57, consisting of two adjacent zinc fingers, in complex with fully methylated DNA at 1.0 Å resolution. The first zinc finger contacts the 5' half (TGC), and the second recognizes the 3' half (CGC) of the recognition sequence. Zfp57 recognizes the two 5-methylcytosines (5mCs) asymmetrically: One involves hydrophobic interactions with Arg178, which also interacts with the neighboring 3' guanine and forms a 5mC-Arg-G interaction, while the other involves a layer of ordered water molecules. Two point mutations in patients with transient neonatal diabetes abolish DNA-binding activity. Zfp57 has reduced binding affinity for unmodified DNA and the oxidative products of 5mC.
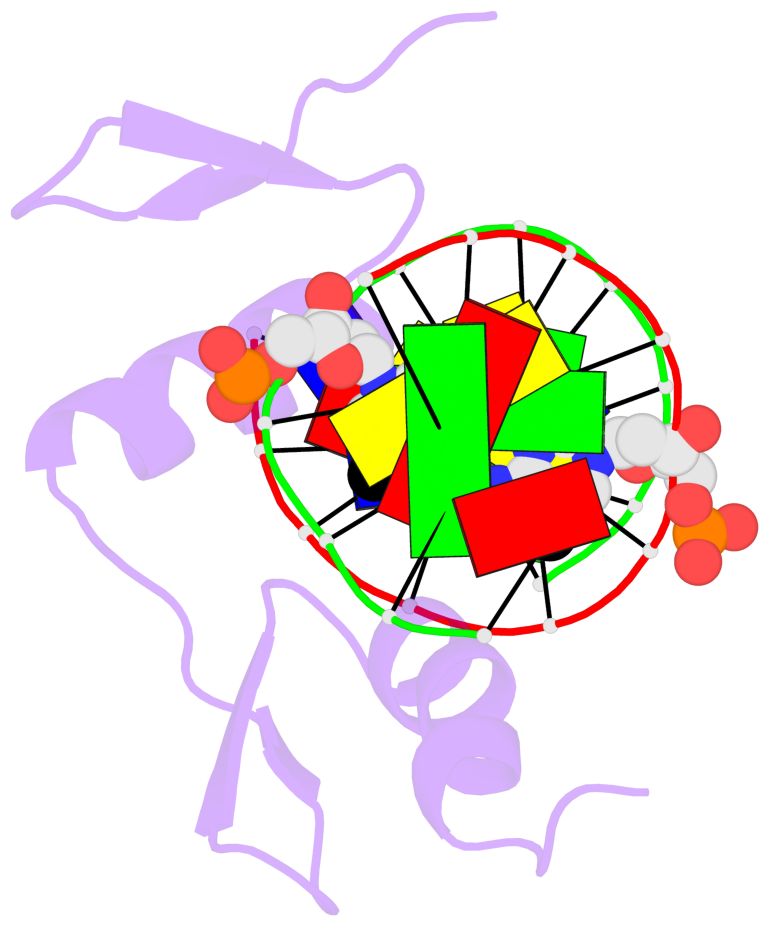
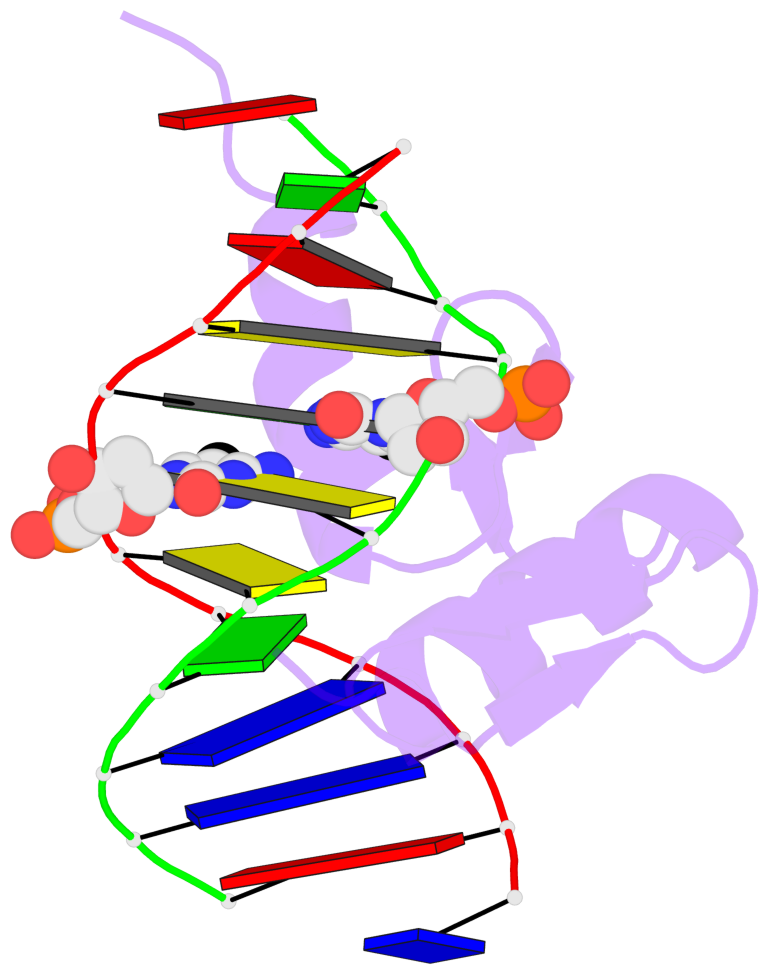
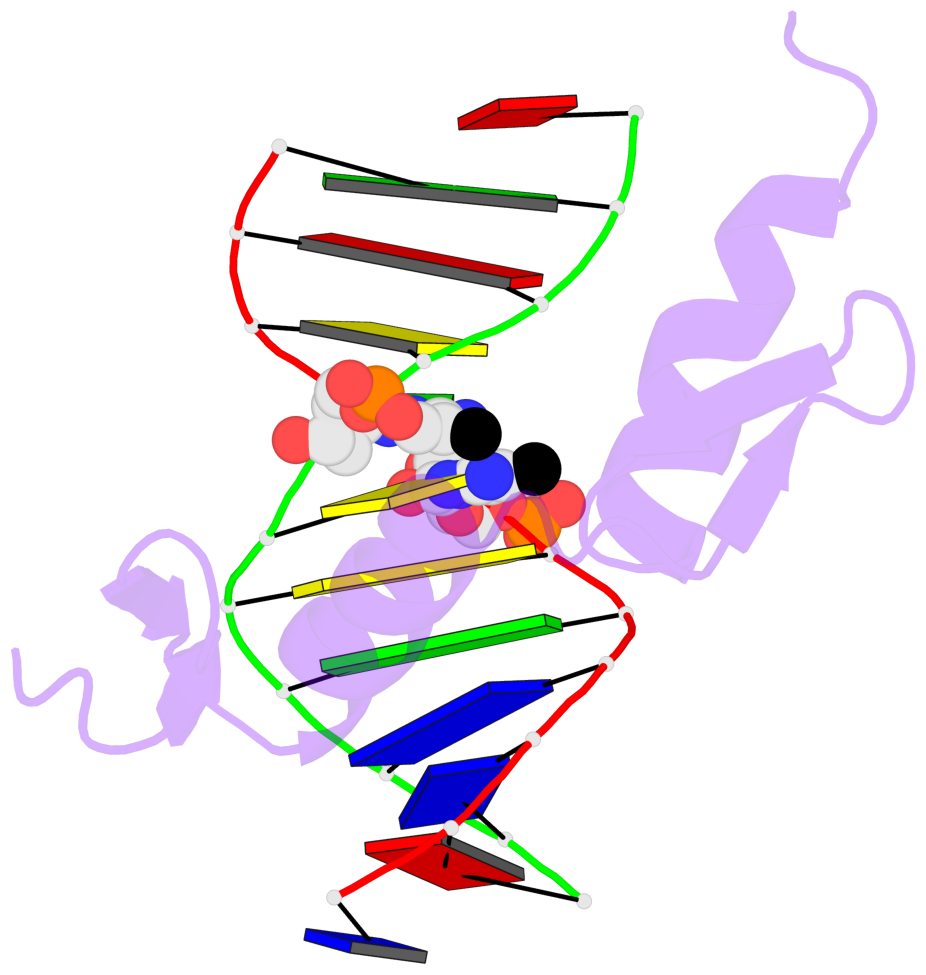
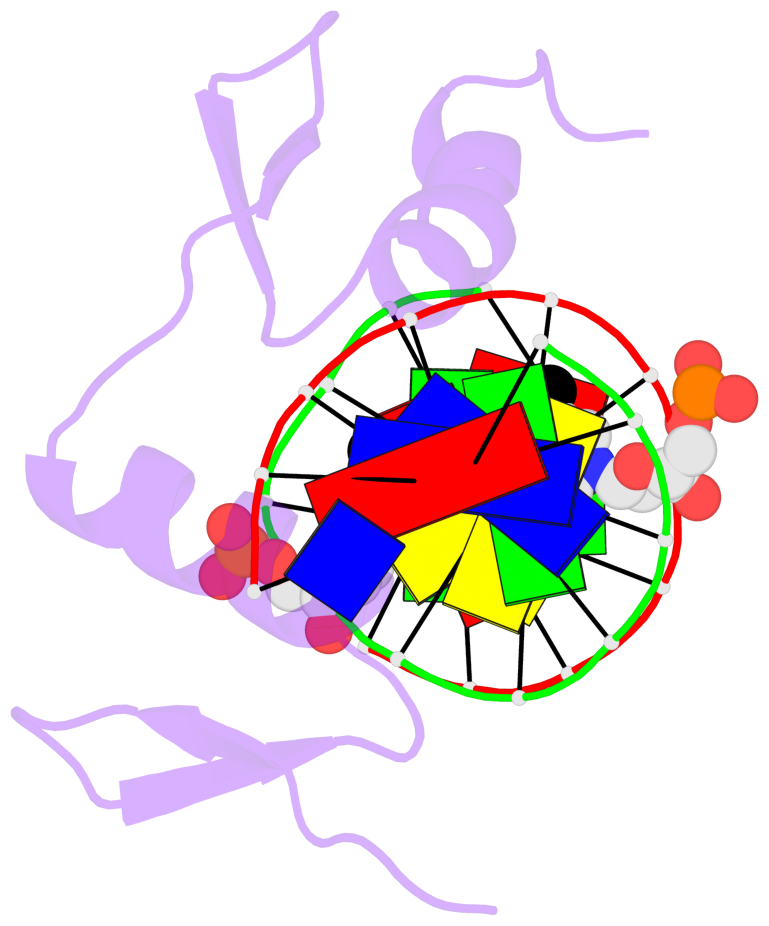
Base-block schematics in six views [summary · contacts · top · homepage · tutorial]
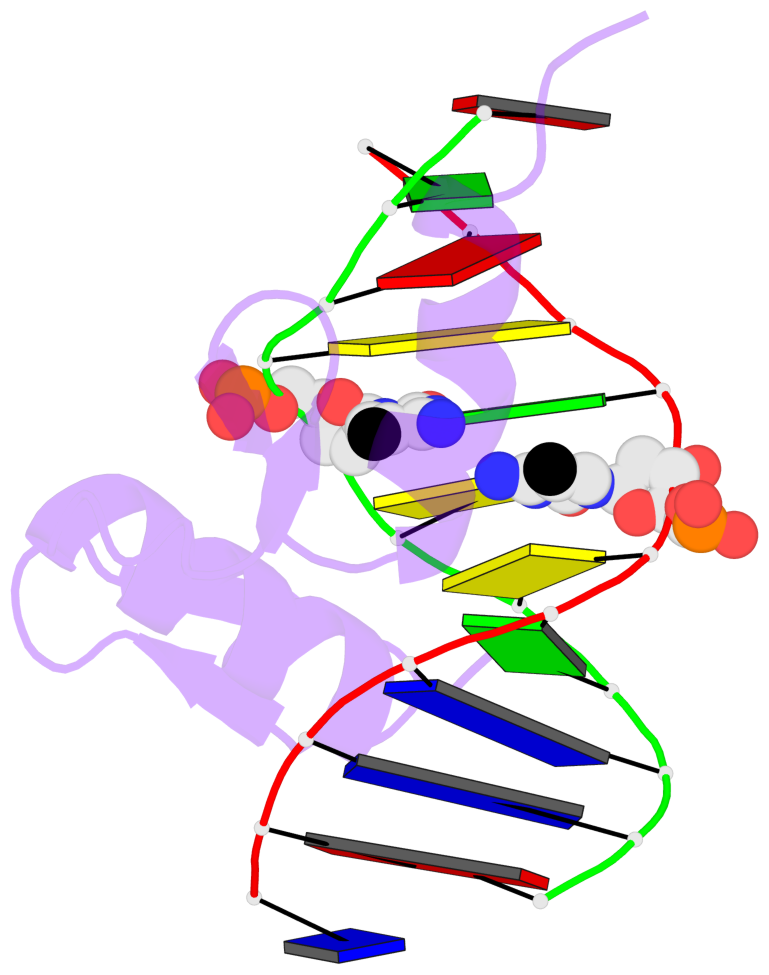
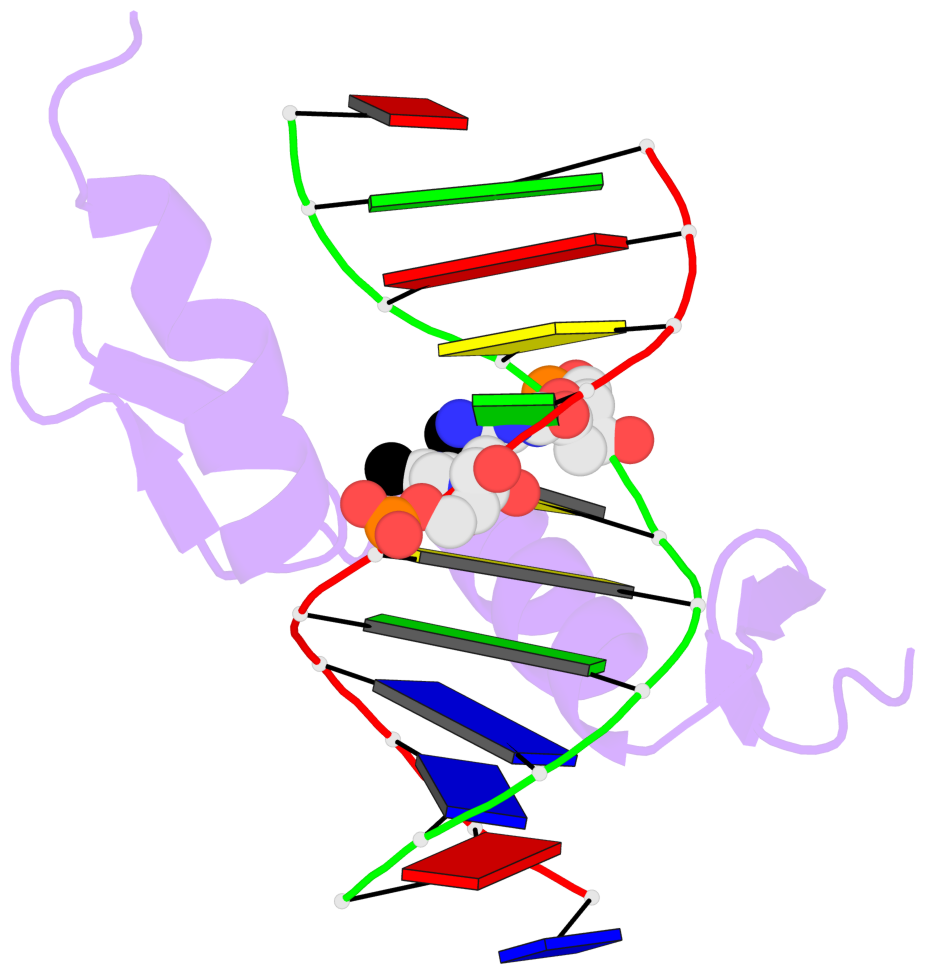
- The 5-methylcytosine group (PDB ligand '5CM') is shown in space-filling model, with the methyl-carbon atom in black.
- Watson-Crick base pairs are represented as long rectangular blocks with the minor-groove edge in black. Color code: A-T red, C-G yellow, G-C green, T-A blue.
- Protein is shown as cartoon in purple. DNA backbones are shown ribbon, colored code by chain identifier.
- The block schematics were created with 3DNA-DSSR, and images were rendered using PyMOL.
- Download the PyMOL session file corresponding to the top-left image in the following panel.
 |
 |
 |
 |
 |
 |
List of 2 5mC-amino acid contacts [summary · schematics · top · homepage · tutorial]
- The contacts include paired nucleotides (mostly a G in G-C pairing), and amino-acids within a 4.5-A distance cutoff to the base atoms of 5mC.
- The structure is oriented in the 'standard' base reference frame of 5mC, allowing for easy comparison and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.