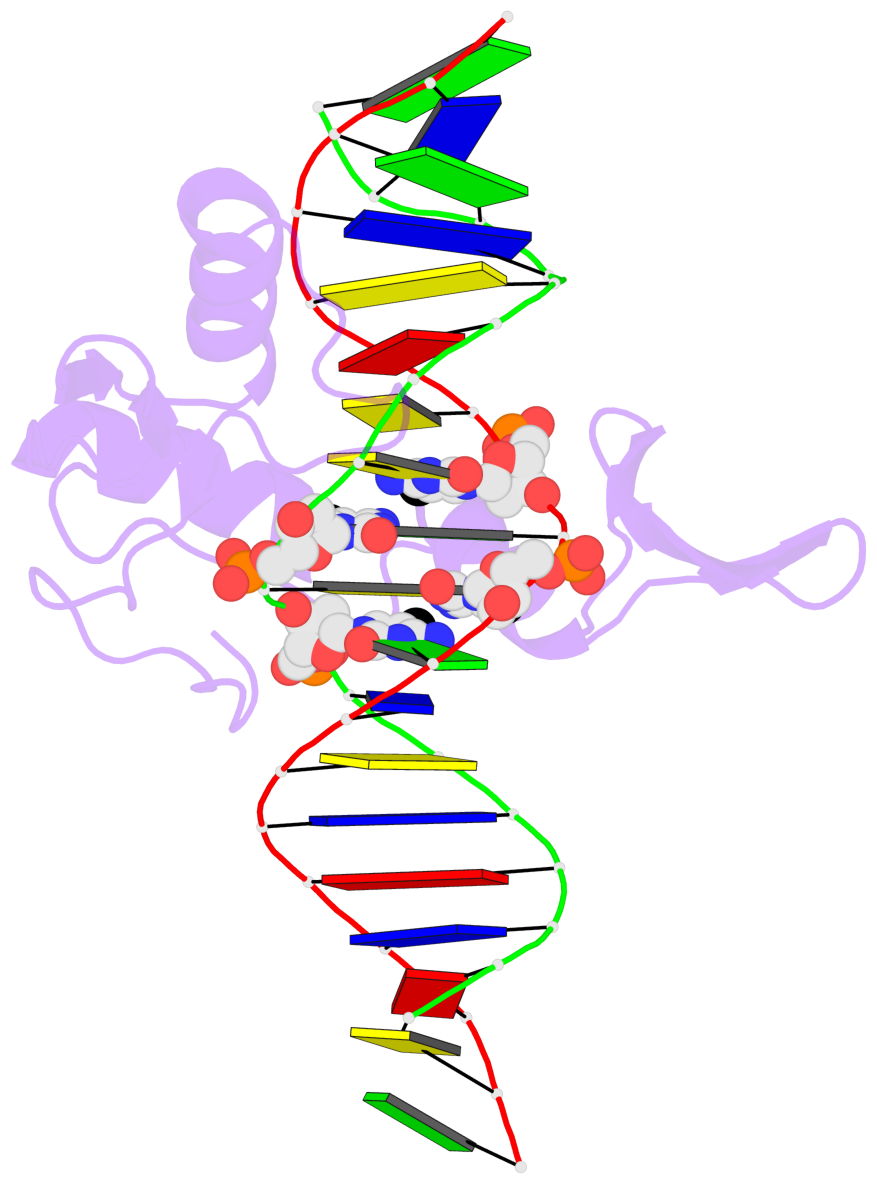
Last updated on 2019-09-30 by Xiang-Jun Lu <xiangjun@x3dna.org>.
The block schematics were created with DSSR and
rendered using PyMOL.
- PDB-id
- 4F6N
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.8 Å)
- Summary
- Crystal structure of kaiso zinc finger DNA binding protein in complex with methylated cpg site DNA
List of 4 5mC-amino acid contacts:
-
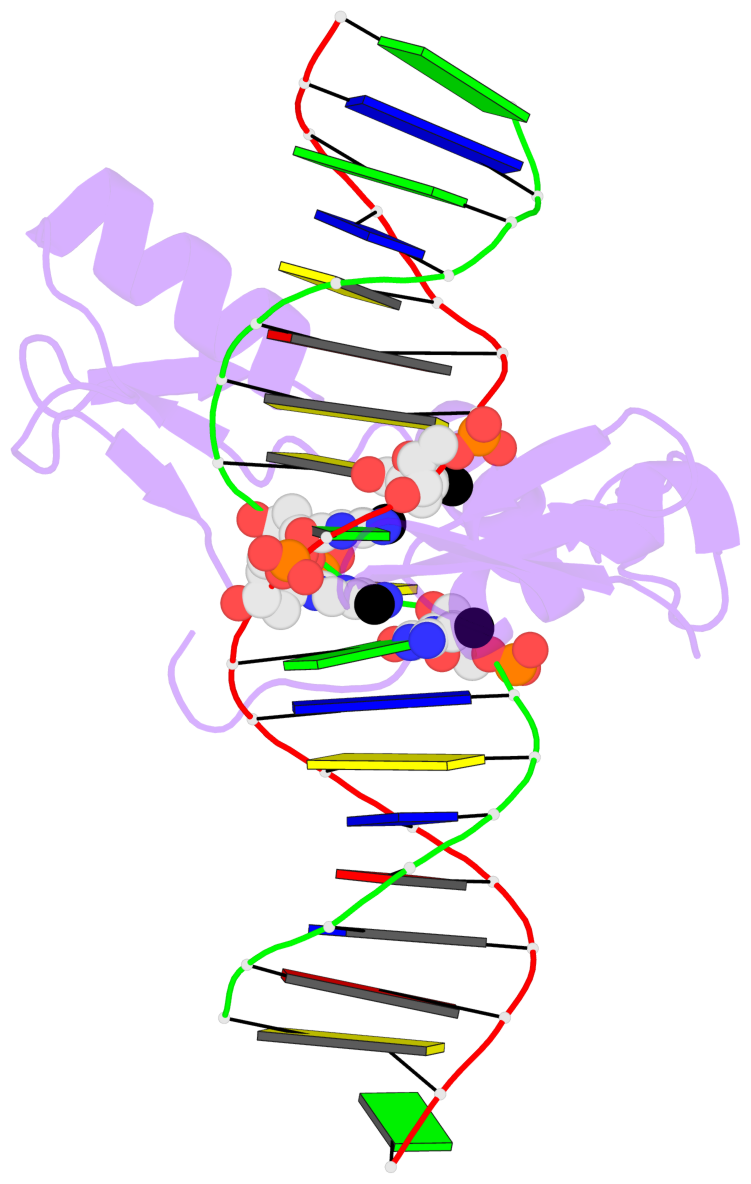
D.5CM8: stacking-with-A.ARG511 is-WC-paired is-in-duplex [+]:CcG/cGG
-
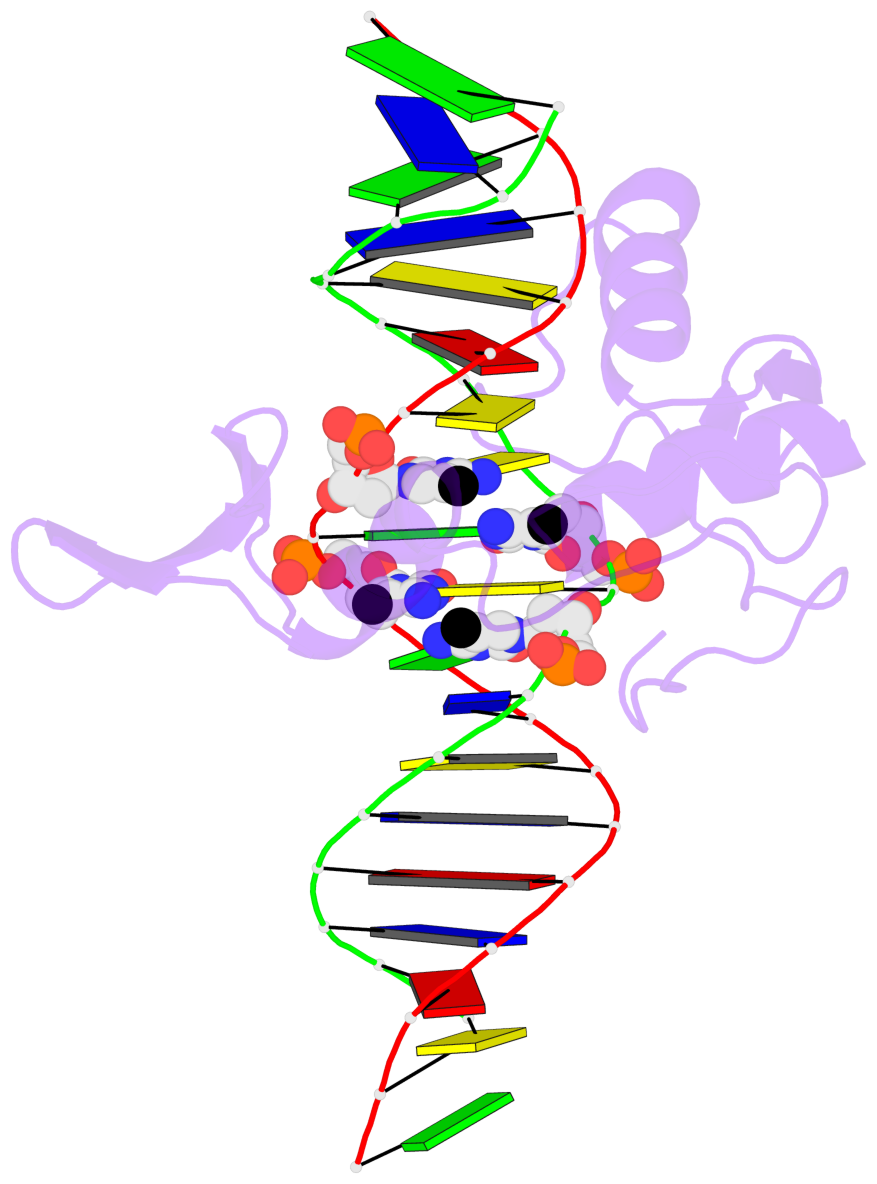
D.5CM10: other-contacts is-WC-paired is-in-duplex [+]:GcG/cGc
-
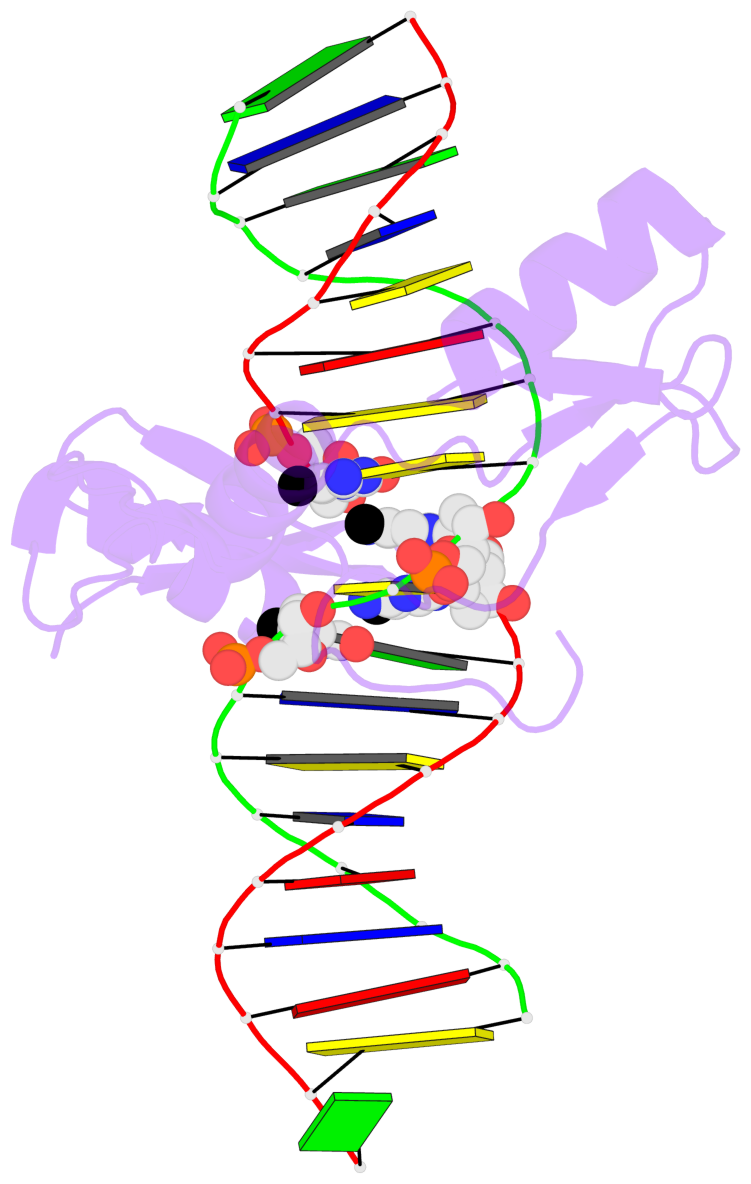
E.5CM28: stacking-with-A.ARG511 is-WC-paired is-in-duplex [-]:cGT/AcG
-
E.5CM30: other-contacts is-WC-paired is-in-duplex [-]:cGc/GcG
direct SNAP output · DNAproDB 2.0
- Reference
- Buck-Koehntop, B.A., Stanfield, R.L., Ekiert, D.C., Martinez-Yamout, M.A., Dyson, H.J., Wilson, I.A., Wright, P.E.: (2012) "Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso." Proc.Natl.Acad.Sci.USA, 109, 15229-15234.
- Abstract
- Methylation of CpG dinucleotides in DNA is a common epigenetic modification in eukaryotes that plays a central role in maintenance of genome stability, gene silencing, genomic imprinting, development, and disease. Kaiso, a bifunctional Cys(2)His(2) zinc finger protein implicated in tumor-cell proliferation, binds to both methylated CpG (mCpG) sites and a specific nonmethylated DNA motif (TCCTGCNA) and represses transcription by recruiting chromatin remodeling corepression machinery to target genes. Here we report structures of the Kaiso zinc finger DNA-binding domain in complex with its nonmethylated, sequence-specific DNA target (KBS) and with a symmetrically methylated DNA sequence derived from the promoter region of E-cadherin. Recognition of specific bases in the major groove of the core KBS and mCpG sites is accomplished through both classical and methyl CH···O hydrogen-bonding interactions with residues in the first two zinc fingers, whereas residues in the C-terminal extension following the third zinc finger bind in the opposing minor groove and are required for high-affinity binding. The C-terminal region is disordered in the free protein and adopts an ordered structure upon binding to DNA. The structures of these Kaiso complexes provide insights into the mechanism by which a zinc finger protein can recognize mCpG sites as well as a specific, nonmethylated regulatory DNA sequence.
- The 5-methylcytosine group (PDB ligand '5CM') is shown in space-filling model,
with the methyl-carbon atom in black.
- Watson-Crick base pairs are represented as long rectangular blocks with the
minor-groove edge in black. Color code: A-T red, C-G yellow, G-C green, T-A blue.
- Protein is shown as cartoon in purple. DNA backbones are shown ribbon, colored code
by chain identifier.
- The block schematics were created with 3DNA-DSSR,
and images were rendered using PyMOL.
- Download the PyMOL session file corresponding to the top-left
image in the following panel.
- The contacts include paired nucleotides (mostly a G in G-C pairing), and
amino-acids within a 4.5-A distance cutoff to the base atoms of 5mC.
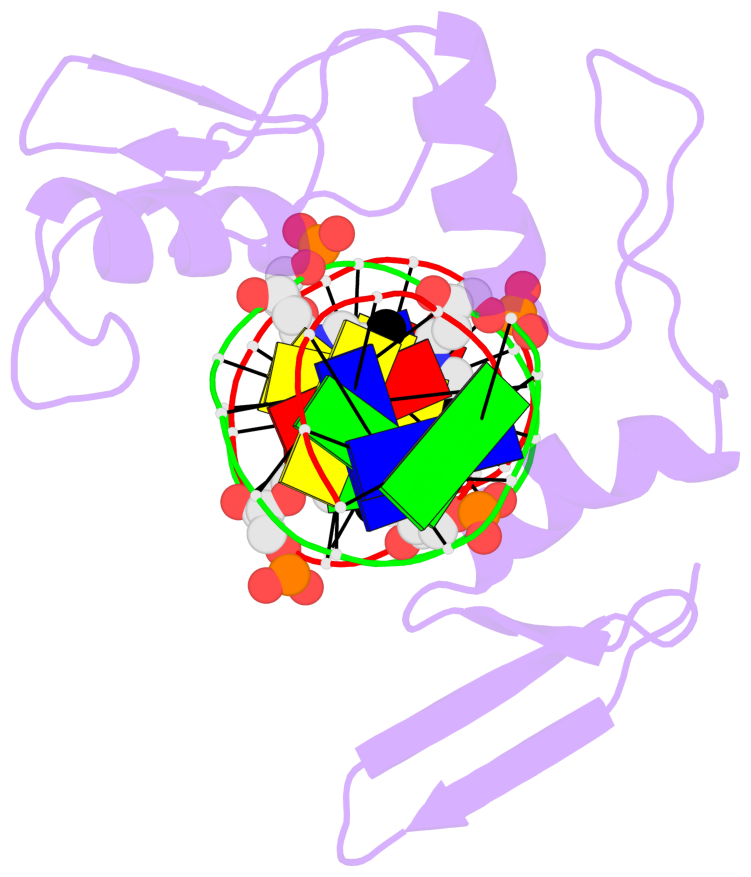
- The structure is oriented in the 'standard' base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
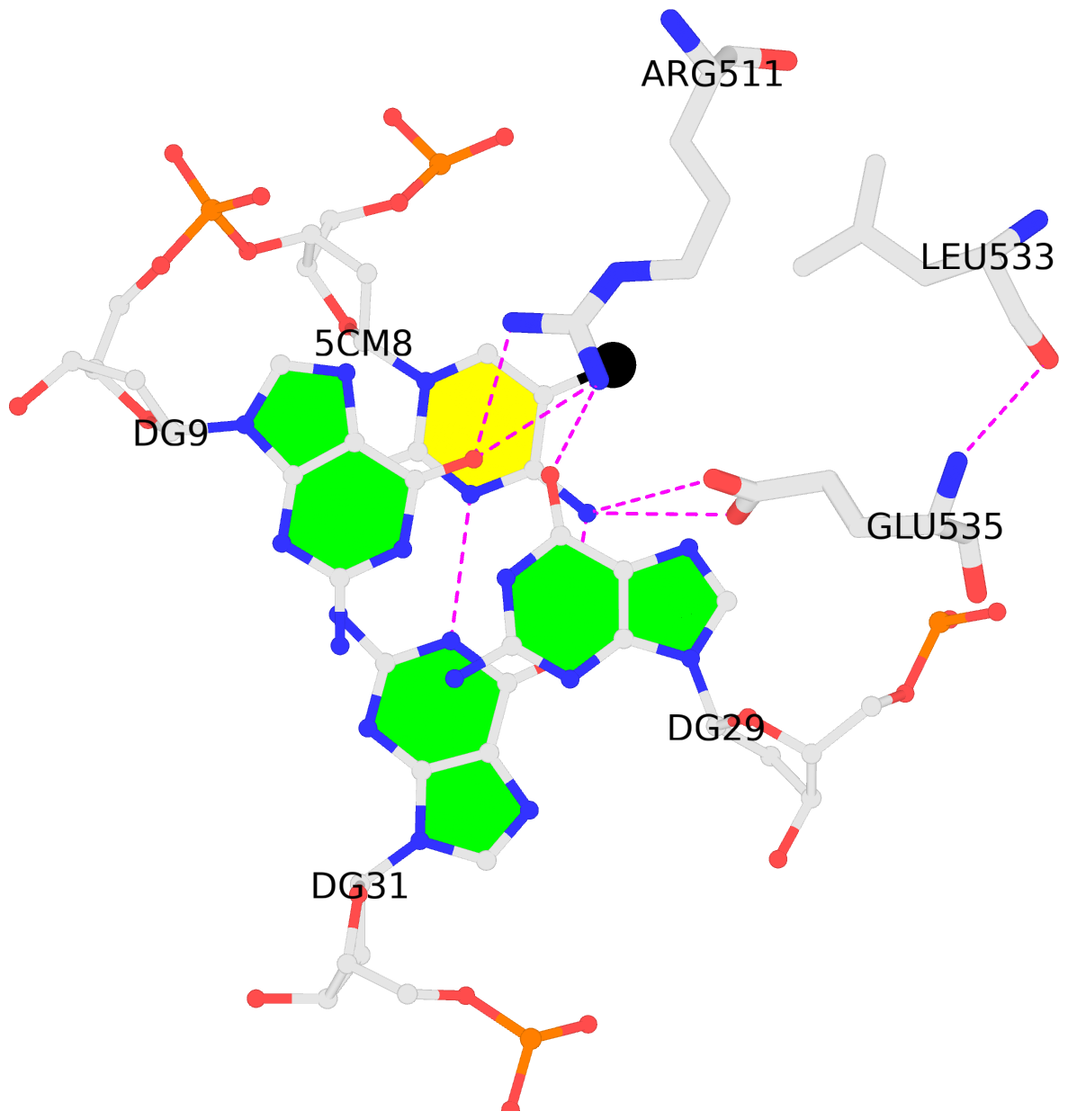 |
No. 1 D.5CM8: download PDB file
for the 5mC entry
stacking-with-A.ARG511 is-WC-paired is-in-duplex [+]:CcG/cGG
|
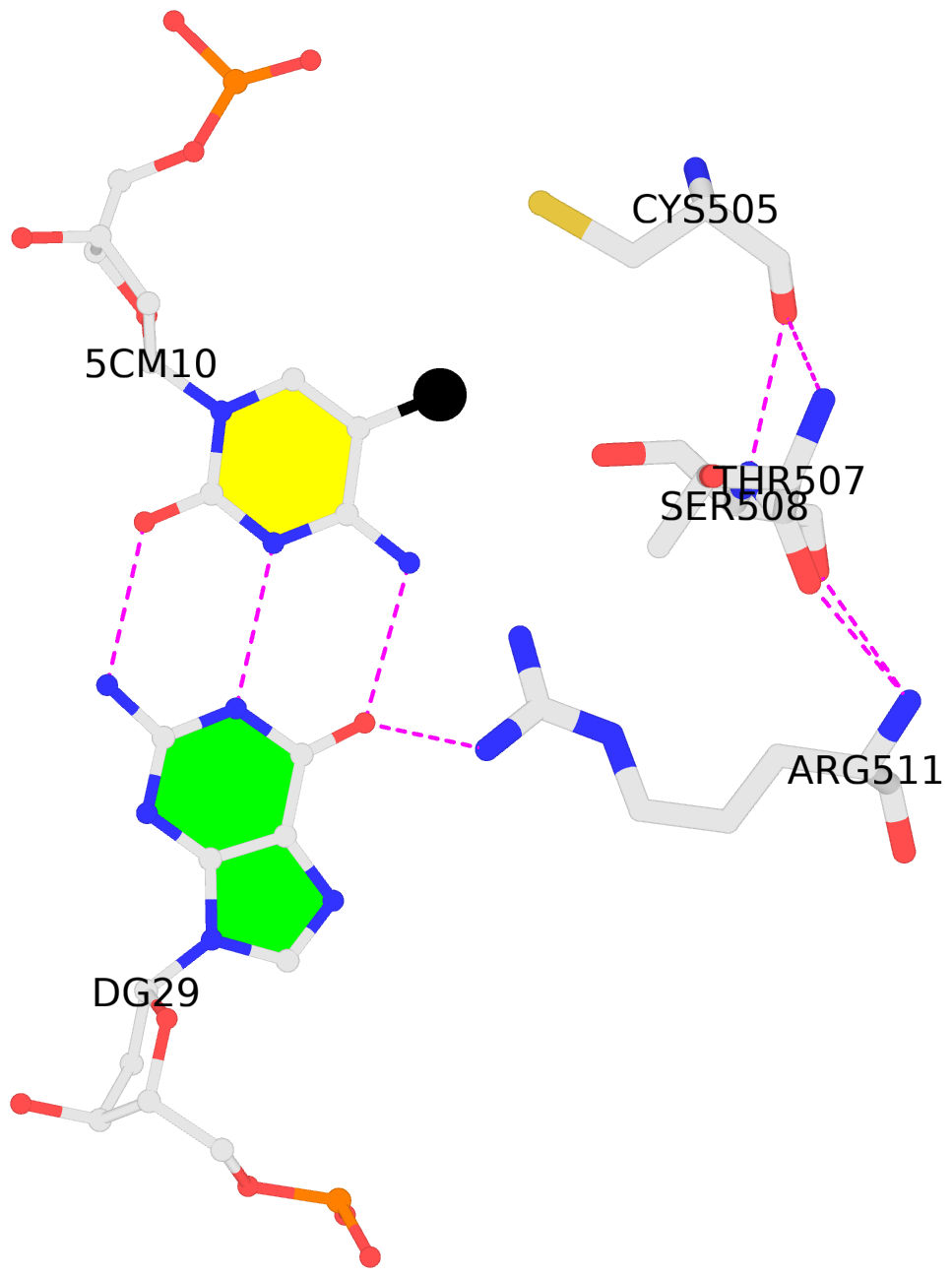 |
No. 2 D.5CM10: download PDB file
for the 5mC entry
other-contacts is-WC-paired is-in-duplex [+]:GcG/cGc
|
 |
No. 3 E.5CM28: download PDB file
for the 5mC entry
stacking-with-A.ARG511 is-WC-paired is-in-duplex [-]:cGT/AcG
|
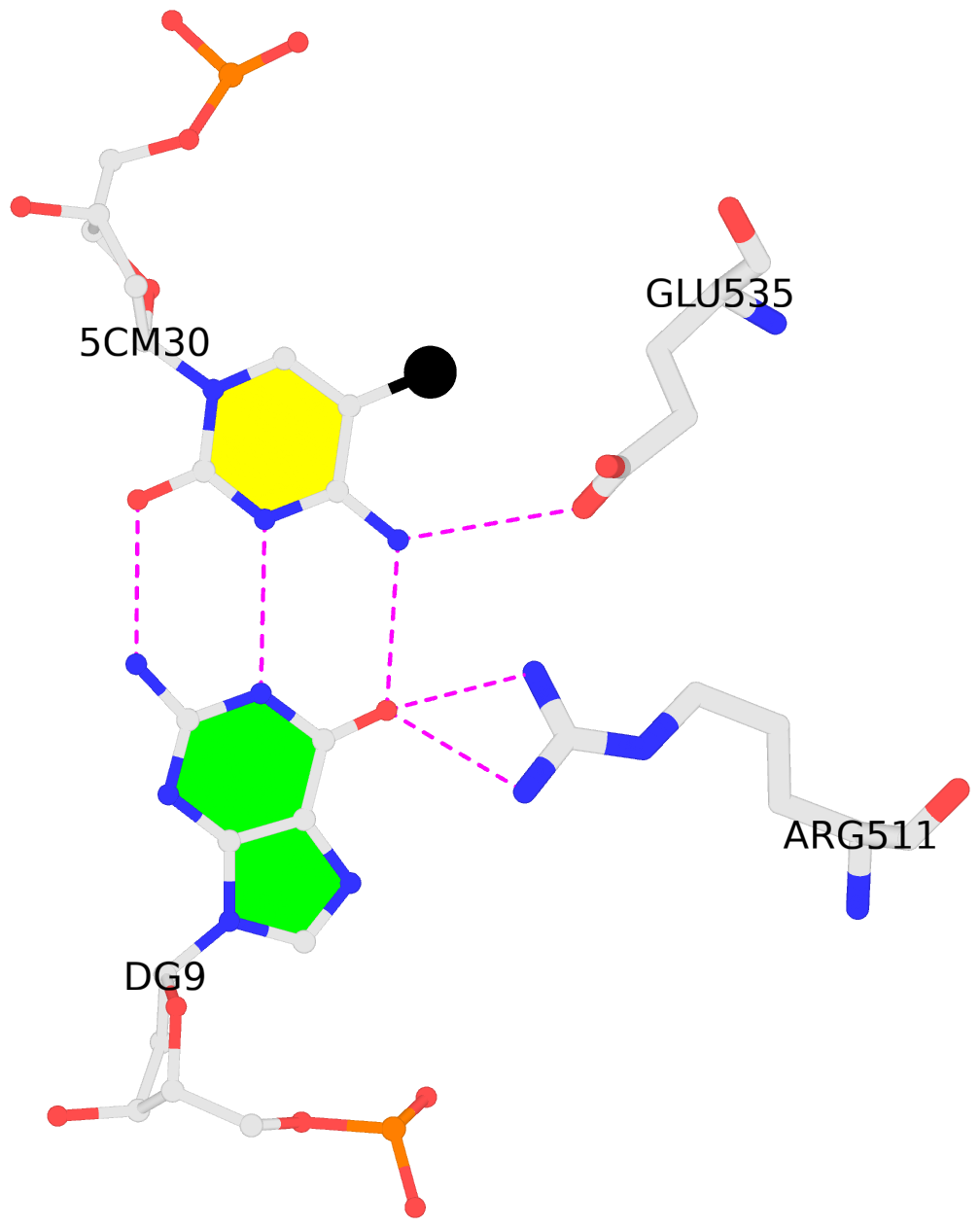 |
No. 4 E.5CM30: download PDB file
for the 5mC entry
other-contacts is-WC-paired is-in-duplex [-]:cGc/GcG
|