5mC interactions in PDB entry 3C2I auto-curated with SNAP
Last updated on 2019-09-30 by Xiang-Jun Lu <xiangjun@x3dna.org>. The block schematics were created with DSSR and rendered using PyMOL.
Summary information and primary citation [schematics · contacts · top · homepage · tutorial]
- PDB-id
- 3C2I
- Class
- transcription regulator
- Method
- X-ray (2.5 Å)
- Summary
- The crystal structure of methyl-cpg binding domain of human mecp2 in complex with a methylated DNA sequence from bdnf
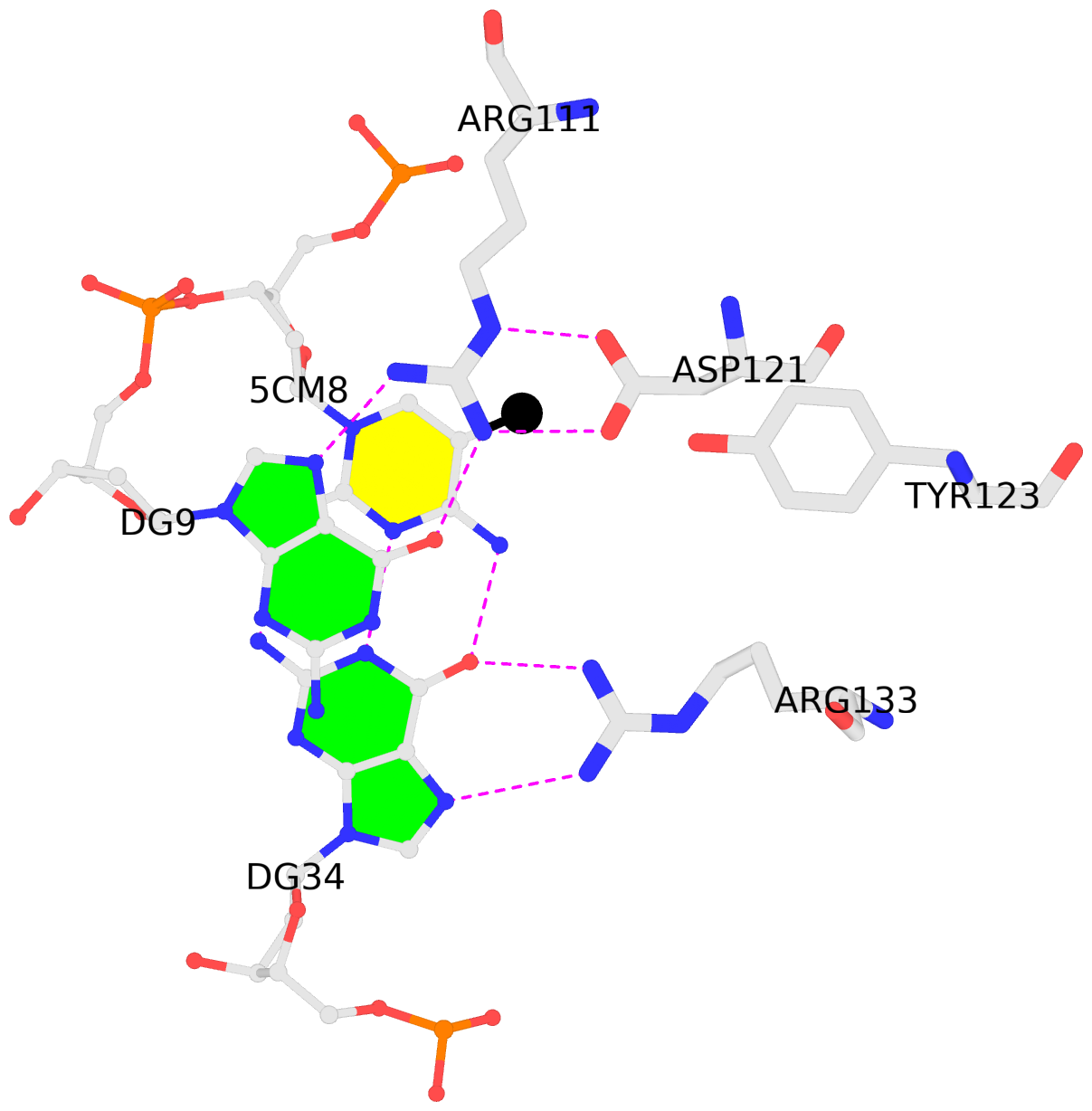
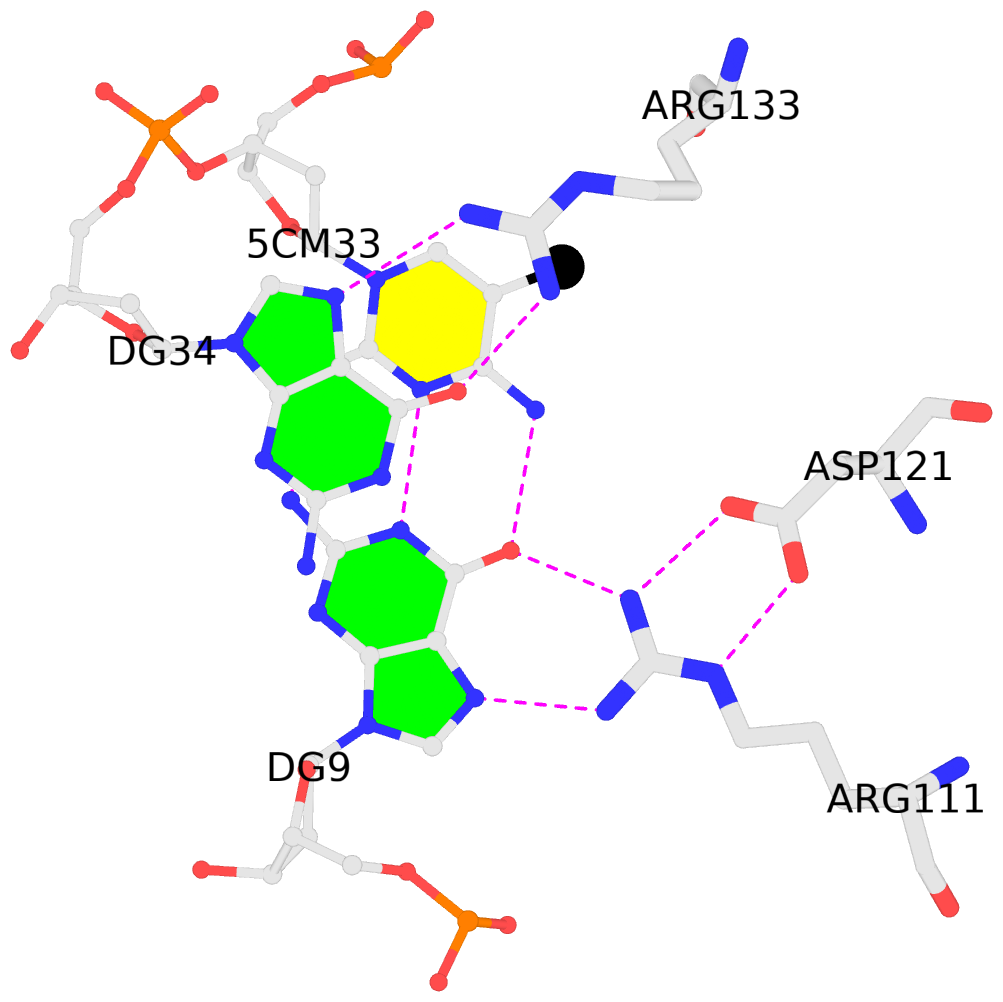
List of 2 5mC-amino acid contacts:-
B.5CM8: stacking-with-A.ARG111 is-WC-paired is-in-duplex [+]:AcG/cGT
-
C.5CM33: stacking-with-A.ARG133 is-WC-paired is-in-duplex [-]:cGG/CcG
-
- Reference
- Ho, K.L., McNae, I.W., Schmiedeberg, L., Klose, R.J., Bird, A.P., Walkinshaw, M.D.: (2008) "MeCP2 binding to DNA depends upon hydration at methyl-CpG." Mol.Cell, 29, 525-531.
- Abstract
- MeCP2 is an essential transcriptional repressor that mediates gene silencing through binding to methylated DNA. Binding specificity has been thought to depend on hydrophobic interactions between cytosine methyl groups and a hydrophobic patch within the methyl-CpG-binding domain (MBD). X-ray analysis of a methylated DNA-MBD cocrystal reveals, however, that the methyl groups make contact with a predominantly hydrophilic surface that includes tightly bound water molecules. This suggests that MeCP2 recognizes hydration of the major groove of methylated DNA rather than cytosine methylation per se. The MeCP2-DNA complex also identifies a unique structural role for T158, the residue most commonly mutated in Rett syndrome.
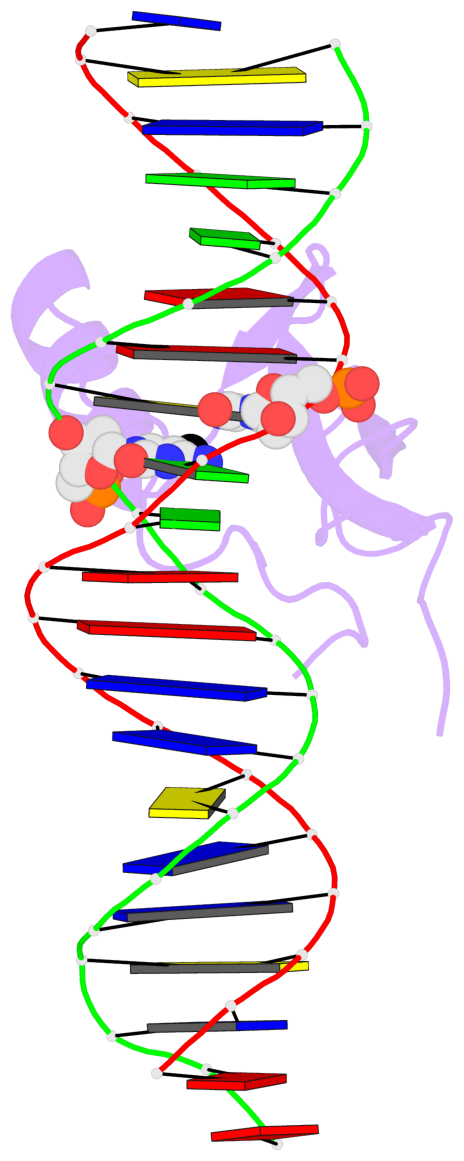
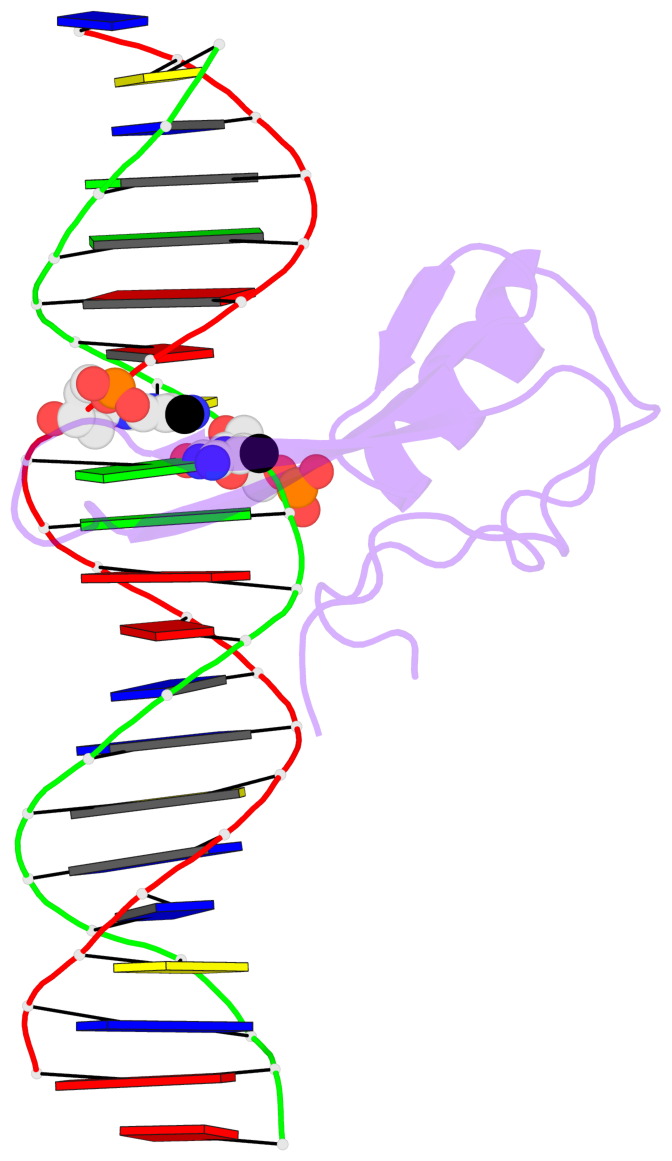
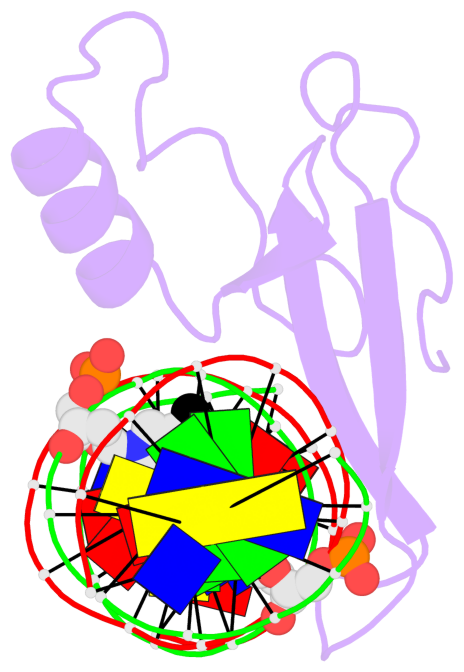
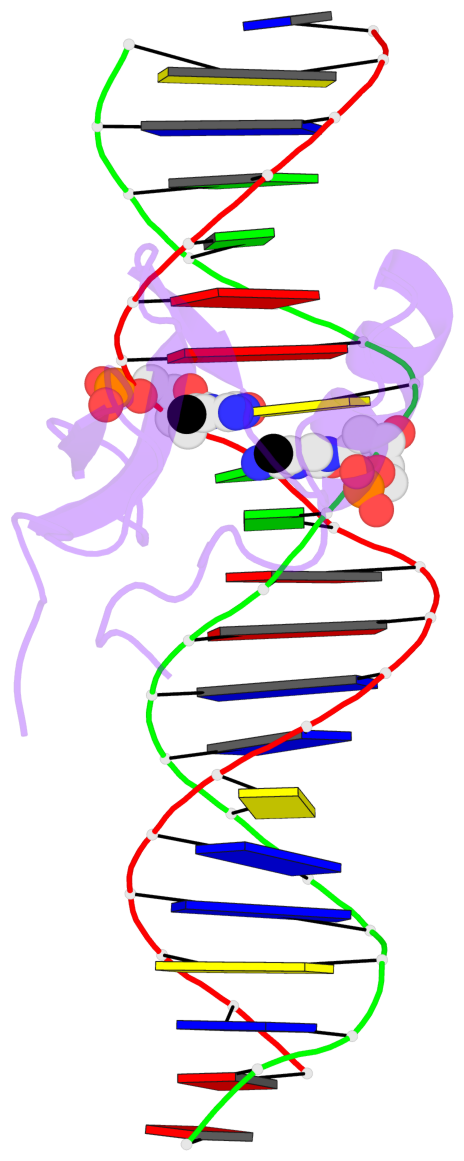
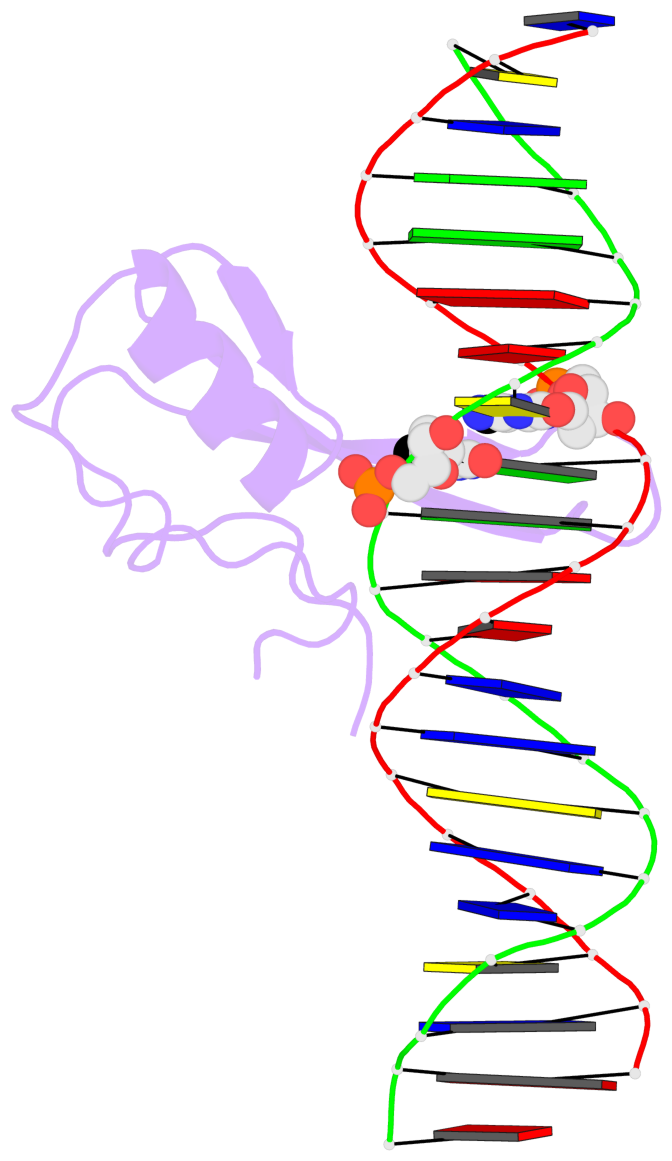
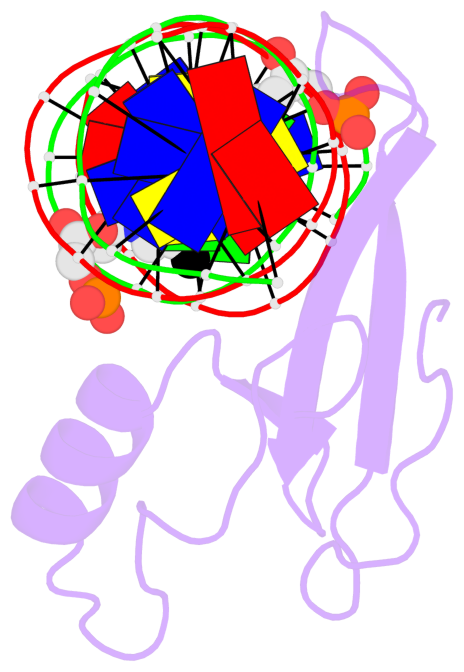
Base-block schematics in six views [summary · contacts · top · homepage · tutorial]
- The 5-methylcytosine group (PDB ligand '5CM') is shown in space-filling model, with the methyl-carbon atom in black.
- Watson-Crick base pairs are represented as long rectangular blocks with the minor-groove edge in black. Color code: A-T red, C-G yellow, G-C green, T-A blue.
- Protein is shown as cartoon in purple. DNA backbones are shown ribbon, colored code by chain identifier.
- The block schematics were created with 3DNA-DSSR, and images were rendered using PyMOL.
- Download the PyMOL session file corresponding to the top-left image in the following panel.
 |
 |
 |
 |
 |
 |
List of 2 5mC-amino acid contacts [summary · schematics · top · homepage · tutorial]
- The contacts include paired nucleotides (mostly a G in G-C pairing), and amino-acids within a 4.5-A distance cutoff to the base atoms of 5mC.
- The structure is oriented in the 'standard' base reference frame of 5mC, allowing for easy comparison and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.